Highlights
-
•
We found 3 major mitochondrial lineages (CRB-G, CRB-PNG & CRB-S in the South Pacific.
-
•
The GBS data provided evidence for gene flow & admixture between different haplotypes.
-
•
We detected high incidence of OrNV infection in all three haplotypes in the region.
-
•
The mitochondrial lineages are not an accurate reflection of the dispersal history.
-
•
The CoxI based method is not a reliable diagnostic marker for phenotypic traits.
Keywords: Coconut Rhinoceros Beetle, Oryctes rhinoceros, OrNV, Nudivirus, the Pacific Islands, Haplotype, Mitochondrial lineage
Abstract
Recently, incursions of the Coconut rhinoceros beetle (CRB), Oryctes rhinoceros, have been detected in south Pacific countries that were previously free of the pest. It has been suggested that this range expansion is related to an O. rhinoceros haplotype that is reported to show reduced susceptibility to the well-established classical biocontrol agent, Oryctes rhinoceros nudivirus (OrNV). We investigated O. rhinoceros population genetics and the OrNV status of specimens collected in Fiji, New Caledonia, Papua New Guinea (PNG), Samoa, Solomon Islands, Tonga, Vanuatu and the Philippines. Based on the sequence of the mitochondrial CoxI gene, we found three major mitochondrial haplotype groups (CRB-G, CRB-PNG and CRB-S) across the region. Haplotype diversity varied between and within countries and a high incidence of OrNV infection was detected in all haplotypes wherever they occurred. The O. rhinoceros population in some countries was monotypic and all individuals tested belonged to a single haplotype group. However, in Samoa we detected CRB-S and CRB-PNG and in Solomon Islands we detected all three haplotype groups. Genotyping-by-Sequencing (GBS) showed genetic differentiation in the O. rhinoceros nuclear genome across populations on different islands and provided evidence for gene flow, resulting in a well-mixed population, despite the presence of different CoxI haplotypes in Solomon Islands. Evidence of admixture was also detected on both islands of Samoa. The current CoxI based method is not a reliable diagnostic marker for phenotypic traits, especially in countries such as Solomon Islands where the mitochondrial haplotypes have come back into sympatry and are mixed. To identify possible mechanisms of resistance to OrNV, further molecular analyses O. rhinoceros in response to virus infection is required. To improve biological control of O. rhinoceros, such analyses will need to be combined with an improved understanding of the population genetics of the pest and the evolutionary history of OrNV in the region.
Graphical Abstract
1. Introduction
The coconut rhinoceros beetle (CRB), Oryctes rhinoceros (L.) (Coleoptera: Scarabaeidae) is a serious pest of coconut and oil palms (see review by Bedford, 2013). In 1909 it was unintentionally introduced to the south Pacific island nation of Samoa from its native Southeast Asia. It has since spread to many parts of the region, causing devastating losses of coconut (Zelazny, 1977a) and oil palm (Ero and Bonneau, 2018). It presents a significant ongoing threat to rural livelihoods in the south Pacific, where these palms are critical to local economies (Ero et al., 2016; Fraenkel, 2010; Tsatsia et al., 2018). In the 1960s, Oryctes rhinoceros nudivirus (OrNV) collected in Malaysia was successfully introduced into Samoa for O. rhinoceros control (Marschal, 1970). Subsequently, it was introduced elsewhere in the region where it also suppressed O. rhinoceros populations, leading to reduced damage to palms (Bedford, 1986; Huger, 2005). Control of O. rhinoceros in the south Pacific by OrNV is widely considered to be a landmark example of classical biological control and it represents one of the few examples of classical biological control involving an entomopathogen (Caltagirone, 1981). Consequently, the system is of considerable ecological and applied significance.
In recent years, new incursions of O. rhinoceros into countries and territories that were previously free of the pest have been reported. The pest was recorded for the first time in Guam in 2007, in Hawaii in 2013 and in Solomon Islands in 2015 (see Marshall et al, 2017). More recently, in 2019, O. rhinoceros was also recorded for the first time in Vanuatu and New Caledonia. Reil et al. (2016) investigated mitochondrial and nuclear DNA gene sequence data from O. rhinoceros collected in Asia, more northerly parts of the Pacific and American Samoa and concluded that two major haplotype groups defined by the Cytochrome C Oxidase subunit I (CoxI) mitochondrial gene correspond to two invasion fronts across the region. One of these haplotype groups was found in Guam, Taiwan, Oahu (Hawaii) and Palau while the other was found in American Samoa, China, Thailand, Vietnam, and Palau. In the investigation, studies of the CAD (carbamoyl-phosphate synthetase, aspartate transcarbamoylase, dihydroorotase) nuclear gene recovered more genetic diversity than studies of the mitochondrial CoxI gene, but the authors concluded that it lacked the resolution required to infer invasion pathways (Reil et al., 2016). Subsequently, it has been suggested that resurgence and the recent spread of the pest in the Pacific is related to an OrNV-tolerant haplotype, “CRB-G”, which is identified by patterns of single nucleotide polymorphisms (snps) in the mitochondrial CoxI gene (Marshall et al., 2017). This haplotype group has previously been identified throughout much of the native and invasive ranges of O. rhinoceros, including Indonesia, the Philippines, Taiwan, Palau, Guam and Solomon Islands (Marshall et al., 2017; Reil et al., 2016).
Although earlier work proposed that O. rhinoceros could evolve resistance to OrNV and that low virulence isolates of virus might occur in different regions where the pathogen has become established (Zelazny and Alfiler, 1991; Zelazny et al., 1992), there is no compelling evidence to support these suggestions (Huger, 2005). OrNV causes chronic infection rather than rapid mortality in its adult hosts, reducing feeding, flight activity, oviposition and mating (Zelazny, 1977a; Zelazny, 1977b). The effects of such chronic infections are difficult to monitor in the field and they do not necessarily cause immediate declines in adult populations. Indeed, studies have shown that OrNV introductions into new islands can decrease adult populations by as little as 10-20% of the original beetle population density (Zelazny et al., 1990) but that the impact of the pathogen can be considerable, especially when it is incorporated into an integrated approach to pest management (Huger, 2005).
We recently determined the full length of the OrNV genomic sequence from an infected O. rhinoceros adult (CRB-G haplotype group) collected in Solomon Islands (Etebari et al., 2020a). The complete circular genome of the virus has 138 amino acid modifications in 53 coding regions when compared with the originally described full genome sequence of the Ma07 strain from Malaysia (NC_011588). Although this demonstrates considerable change in the pathogen, further sequence analyses of more OrNV isolates and investigation of their effects on host insects from multiple populations is needed to determine if such mutations compromise the efficacy of OrNV as a biocontrol agent. The origin of the OrNV that we found infecting O. rhinoceros in Solomon Islands is not known. It is possible that it was accidentally released from laboratories in Honiara that conducted OrNV bioassays there between 2016 and 2018, but there is no evidence for this. Analysis of OrNV genomic structural and transcriptional variation in samples collected across the Pacific and the Philippines showed that OrNV strains from Solomon Islands and the Philippines are closely related, while those from PNG and Fiji formed a distinct adjacent clade (Etebari et al., 2020b).
In their report that the ‘CRB-G’ haplotype group is associated with increased tolerance to OrNV, Marshall et al. (2017) also describe two further mitochondrial haplotype groups. One of these is predominantly from Papua New Guinea and is more divergent from the ‘CRB-G’ clade than the remaining haplotype group; both these haplotype groups are referred as ‘CRB-S’ (susceptible to OrNV) by Marshall et al. (2017). Using many nuclear snp markers (approximately 7,500 with 75% missing data), Reil et. al, (2018) were able to resolve the likely movement pathways of O. rhinoceros through the region with more clarity than they were able to do using the CAD nuclear gene (Reil et al., 2018). Further, they used the single mitochondrial snp test developed by Marshall et al. (2017) to distinguish between the ‘CRB-G’ and ‘CRB-S’ haplotypes. In the analysis of the snp data they found evidence for geneflow between the ‘CRB-G’ and ‘CRB-S’ mitochondrial groups in Palau (Reil et al., 2018), supporting previous evidence for such admixture provided by the CAD gene sequence data (Reil et al., 2016),. Consequently, the authors note that genetic admixture between haplotypes compromises the utility of the current CoxI based assay and suggest that more reliable and sophisticated methods need to be developed.
In this study, we investigated the population genetics of O. rhinoceros sampled from across the islands of the south Pacific. We combined analysis of the mitochondrial CoxI gene with ∼6,500 snp markers obtained from the nuclear genome using a genotyping‐by‐sequencing (GBS) method. Our study provided the first GBS data for O. rhinoceros specimens collected from Fiji, Samoa, PNG, Tonga and Solomon Islands. We investigated the geographical distributions of the different haplotype groups and gene flow across different islands and between the mitochondrial haplotypes. We then tested for a relationship between different mitochondrial haplotypes and the incidence of OrNV in the gut tissue of field collected beetles as detected by PCR.
2. Materials and Methods
2.1. Insect Collection
Adult O. rhinoceros were collected in traps baited with aggregation pheromone (Oryctalure, P046-Lure, ChemTica Internacional, S. A., Heredia, Costa Rica) lures. Collections were made in Fiji (Viti Levu and Vanua Levu), New Caledonia (Nouméa), Papua New Guinea (Kimbe, New Britain), Samoa (Upolu and Savai'i), Solomon Islands (Guadalcanal, Russell Islands, Gizo, Kolombangara and Santa Cruz Islands), Tonga (Tongatapu), and Vanuatu (Efate) in the South west Pacific and in the Philippines (Los Banos) between January and October, 2019. Upon removal from traps, individual beetles were preserved in 95% ethanol in separate containers and shipped to the University of Queensland, Brisbane, Australia for further analysis.
2.2. DNA extraction and mitochondrial CoxI gene analysis
To characterise the different mitochondrial DNA haplotypes in the O. rhinoceros samples from across the region, we sequenced the CoxI gene in 260 individuals. We also included previously sequenced O. rhinoceros CoxI gene sequences from other geographical regions, which are publicly available on NCBI, in our phylogenetics analysis.
DNA was extracted from a leg of adult O. rhinoceros using a DIY spin column protocol using 96-well silica filter plates from Epoch life sciences (Missouri City, TX, USA) (Ridley et al., 2016). A small fragment of the CoxI gene was amplified in these individuals with the primers LCO1490 and HCO2198 (Folmer et al., 1994). PCRs were carried out as 25 µl reactions containing 0.03 units of MyTaq™ (Bioline), 1 × buffer and 0.2 μM of forward and reverse primers and 2 μl of template DNA. The PCR conditions were as follows: 95°C for 3 min, 40 cycles consisting of denaturation at 95°C for 15s, annealing at 55°C for 30s and 72°C for 15s. PCR products were cleaned using one unit each of Exonuclease I and Antarctic Phosphatase (New England Biolabs, Massachusetts, USA), with sequencing reactions performed using an ABI3730 Genetic Analyzer (Applied Biosystems) at Macrogen Inc. Seoul, South Korea. PCR products were sequenced bi-directionally after confirmation by 1% agarose gel electrophoresis. After primer trimming and alignment, these two CoxI regions overlapped by 621 bp and this segment was used for further phylogenetic analysis. For the construction of the maximum-likelihood phylogenetic tree, we selected a nucleotide substitution model based on the consensus outcome of a hierarchical likelihood ratio test (hLRT), Bayesian information criterion (BIC) and Akaike information criterion (AIC). For the mitochondrial CoxI phylogenetic inference, the General Time Reversible (GTR) +G rate variation, +T topology variation was selected from all tests using jModelTest 2.1.10 (Darriba et al., 2012). The maximum-likelihood phylogeny was then constructed using CLC Genomics Workbench (v 20) starting tree construction method under the nucleotide-substitution models mentioned above with 1,000 bootstraps. We also used the GTR+G model to construct a Bayesian phylogeny using mrBayes (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) with eight chains (each 11,000,000 iterations) and a burning length of 100,000. The haplotype network was generated based on 621 bp of the CoxI gene region from a total of 260 individuals using PopART 1.7.2 (Leigh and Bryant, 2015).
2.3. Genotyping-by-Sequencing analysis
To assess gene flow across the nuclear genome of these populations we generated genome-wide snp data using a genotyping-by-sequencing method. We followed the protocol and adaptor regime described by Hereward et al., (2020), which is based on the methods of Elshire et al. (2011), Poland et al. (2012) and Peterson et al. (2012), with barcodes based on Caporaso et al. (2012). Briefly, samples were normalised to 200ng and double-digested with MspI and PstI. We ligated 96 unique forward barcodes, and three different plates of reverse barcodes so that every sample had a unique forward and reverse barcode combination. Each plate of 96 samples was then pooled and size selected on a Blue Pippin (Sage Science) to 300-400bp. Each of these pools was PCR amplified to complete the adaptors and add indices. We then normalised and pooled these PCR products for a total of 198 individuals and sequenced the libraries on 2/3 of a lane with PE150 Illumina sequencing at Novogene (Beijing, China).
The sequence data were demultiplexed, assembled, and snps called using Stacks (version 2.55) using the options -m4 -M3 and n3 (Catchen et al., 2013). We produced two variant call (vcf) files using the populations command in Stacks and applying a heterozygosity threshold of 0.65, one of these had multiple snps per locus (and was used to construct the PCA), and one was limited to one snp per locus (for the structure analysis). We then filtered the two vcf files using vcftools, with the data first being filtered to a minor allele count of three (one heterozygote and one homozygote). This allows the conservative removal of singleton snps that are likely to be errors, without discarding rare alleles (Linck and Battey, 2019). We set a minimum depth of 5, and kept only biallelic snps. We filtered the data for missing data in three steps. First, any marker missing more than 50% data was discarded (i.e. 50% of individuals were not genotyped at that marker), to remove the markers most affected by missing data. Second, any individual that had missing data at more than 70% of the markers was discarded, to remove the individuals that had poor quality genotyping, we retained 185 out of 198 samples. Finally, any marker missing more than 5% data was discarded to produce a filtered dataset with relatively little missing data (∼3%). We plotted a PCA (Principal Component Analysis) using the adegenet package in R (Jombart, 2008), based on 6,561 snps. We then ran structure (Pritchard et al., 2000) using a filtered dataset with 1,138 snps. Structure is an individual-based clustering algorithm that assigns individuals to each of K population clusters using Hardy-Weinberg equilibrium and linkage information in genetic markers. We ran seven runs of structure for each value of K from 1 to 10, each of these runs used the admixture model, was run for 1 million iterations with 100,000 iterations to burn in. The results for multiple runs were then permuted and plotted using Clumpak server (Kopelman et al. 2015), and the most likely value for K was inferred using the Evanno et al (2005) DeltaK method implemented in Structure Harvester (Earl and vonHoldt, 2012)
2.4. Incidence of OrNV infection in adult O. rhinoceros
To determine the incidence of OrNV in O. rhinoceros specimens collected in the field from across the Pacific, the gut tissue was carefully dissected from adult beetles and total DNA from individual specimens was extracted as described previously. The presence of OrNV in gut tissue was confirmed by PCR amplification of a 945 bp product using the OrV15 primers that target the OrNV-gp083 gene (Richards et al., 1999). The PCR product was visualized by gel electrophoresis and validated by Sanger sequencing.
3. Results
3.1. Mitochondrial CoxI analysis
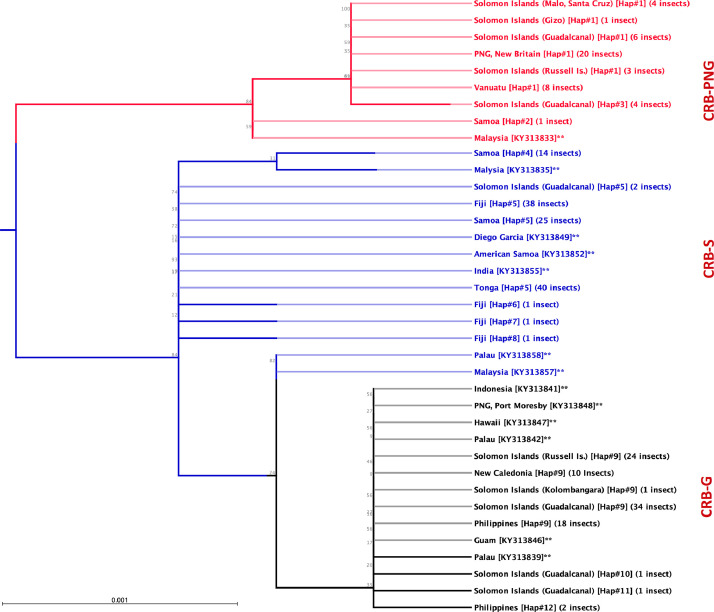
Twelve different haplotypes, each belonging to one of three major haplotype groups (CRB-S, CRB-G and CRB-PNG), were identified in field-collected O. rhinoceros (Fig. 1 and Supplementary Fig S1). These sequences have been submitted to NCBI under GenBank accession numbers MN809502-MN809525.
Fig. 1.
The Maximum-Likelihood phylogeny of O. rhinoceros based on partial CoxI gene sequence (621bp). 12 different haplotypes of O. rhinoceros, in three major haplotype clades (CRB-PNG, CRB-S and CRB-G) were identified. The tree constructed under the nucleotide-substitution model GTR+G+T with 1,000 bootstraps. The CoxI sequence of individuals highlighted with ** previously sequenced by other researchers and downloaded from NCBI and the number in brackets represents their NCBI accession code.
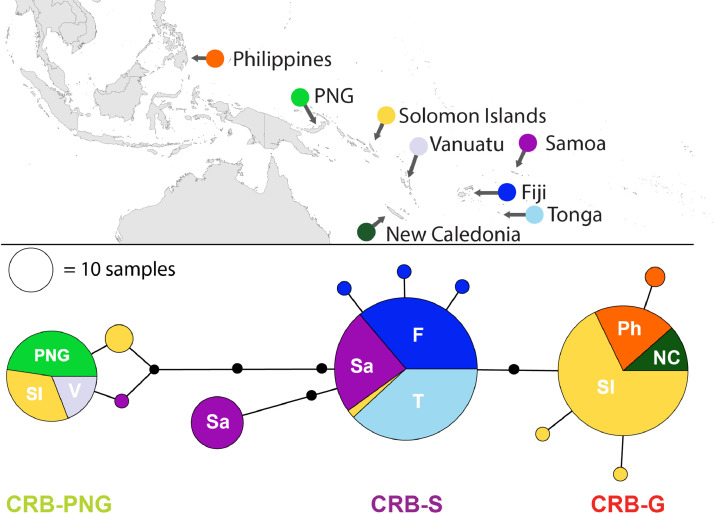
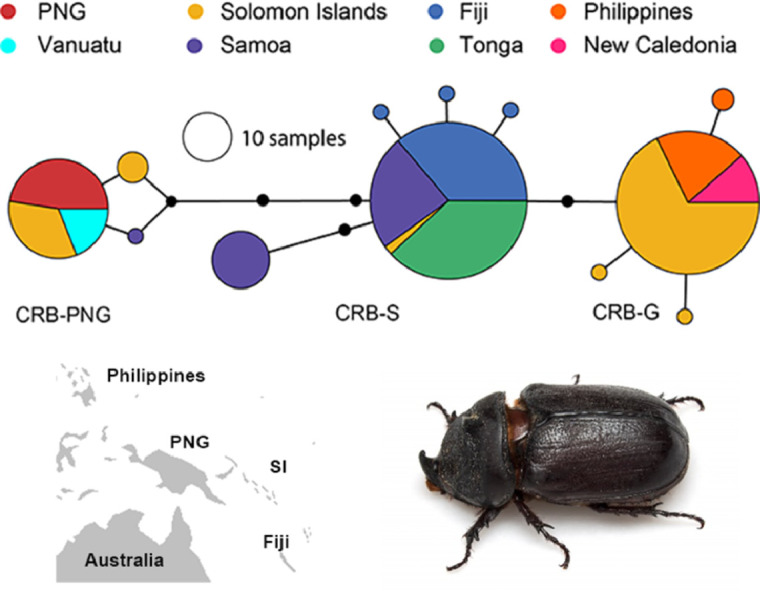
Haplotype diversity varied considerably between regions, and between and within countries. In Tonga, the population was completely monotypic and all individuals tested belonged to a single mitochondrial haplotype (CRB-S; Fig. 1). The population in Fiji contained four different haplotypes, all of which also belonged to the CRB-S haplotype groups (Fig. 1). In samples from Samoa, three distinct haplotypes were identified; two of these were more frequent in the population and both belonged to the CRB-S haplotype groups (Samoa Hap#4, and Hap#5; 14 and 24 individual beetles respectively). A third haplotype, which differed from the other haplotypes in Samoa by 4 to 6 base pairs (Figs.1 and 2; Table S1), belonged to the CRB-PNG haplotype group (Samoa Hap#2; only one individual collected from Upolu). The haplotype network generated for the CoxI gene suggests that O. rhinoceros from Tonga and Fiji, and most of those collected from Samoa, are almost identical with low mitochondrial genetic variation between these individuals (Fig. 2 and Table S1).
Fig. 2.
O. rhinoceros CoxI haplotype network. The network is based on 621 bp of the CoxI gene region from a total of 260 individuals collected from Pacific Islands (created in PopART (Leigh and Bryant, 2015)). The circles represent haplotypes and their diameters are proportional to relative abundance across all samples in the study (black circles along branches represent single nucleotide differences). The different colours show the distribution of the different haplotypes between the different countries.
All O. rhinoceros individuals collected from PNG (Kimbe, New Britain) belonged to the CRB-PNG haplotype group and specimens conforming to this clade were also identified in samples from Vanuatu and Solomon Islands (Malo, Santa Cruz Islands; Gizo; Guadalcanal and Russell Islands) (Figs. 1 and 2; Table S1). Specimens with a high degree of similarity to this haplotype group were also identified from Samoa, Guadalcanal and previously sequenced individual collected from Malaysia (Figs. 1 and 2). This haplotype group showed maximum polymorphism (5-8 nucleotide differences) compared to other haplotypes collected from the South Pacific islands and the Philippines (Table S1).
Four haplotypes conforming to the haplotype group previously described as CRB-G were detected in specimens collected in Solomon Islands (Guadalcanal, Russell Islands and Kolombangara), New Caledonia, and in the Philippines (Figs. 1 and 2; Table S1). This group of beetles showed more similarity to the CRB-S than to the CRB-PNG haplotype group based on the number of snps in their CoxI gene (Table S1). Previously sequenced specimens from Guam, Hawaii, Palau and PNG (Port Moresby) were also classified under this haplotype group (See Fig. 1 and Fig S1). The presence of more than one mitochondrial haplotype group was detected in Solomon Islands, PNG, Palau, Samoa and Malaysia.
3.2. Genotyping-by-Sequencing analysis
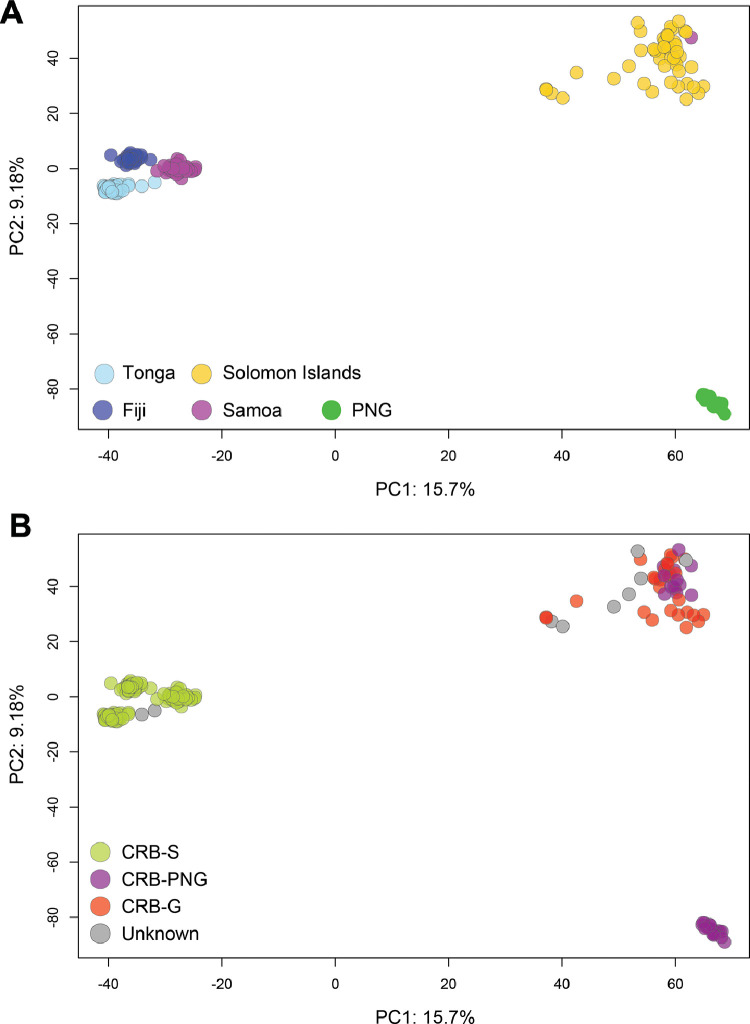
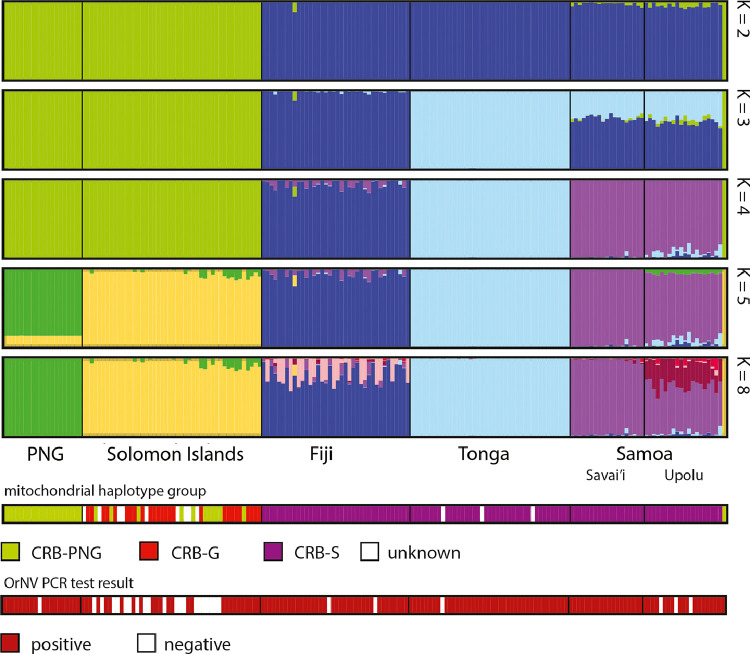
Despite the presence of both the CRB-G and CRB-PNG CoxI haplotypes in Solomon Islands, our population genetic analysis of nuclear DNA data shows that the O. rhinoceros population there is well-mixed in genetic terms. This is supported by the PCA plot (Fig 3) and the structure analyses. Solomon Islands is closely related to PNG and is assigned to the same cluster when K=4. It is only separated out in higher values of K (Fig. 4). The beetles collected from PNG clustered close together in the PCA (Fig. 3A) and are distinct from those collected in Solomon Islands in the PCA, despite the admixture evident between the CRB-PNG and CRB-G CoxI haplotype groups there (Fig. 4). Only two individuals from the CRB-S haplotype group were collected from Guadalcanal in this study and we were unable to include them in our GBS analysis at the time of investigation.
Fig. 3.
Principal component analysis (PCA) plots of the GBS data coloured by country of origin (A), and by CoxI haplotype groups (B). We produced a variant call file (vcf) using the populations command in stacks and applying a heterozygosity threshold of 0.65 with multiple snps per locus. We plotted a PCA using the adegenet package in R (Jombart, 2008), based on 6,561 snps. We couldn't confirm the CoxI haplotypes of some individuals due to low quality of reads from sanger sequencing and marked them as “unknown” in PCA plot.
Fig. 4.
Results of multiple analyses using the individual-based Bayesian clustering program structure. Analyses were conducted with different values of K (the number of hypothetical populations the program should sort individuals into). Plots are shown for K=2, 3, 4, 5 and 8 (see supplementary data for others), each vertical bar in the plot represents one individual and the colouring of the bars represents the posterior probability of that individual belonging to each of the K hypothetical populations based on Hardy-Weinberg equilibrium and linkage. At the bottom of the plots, the mitochondrial haplotype group of that individual beetle is shown (CRB-G, CRB-S and CRB-PNG). The OrNV infection status also showed for each individual.
Specimens from Tonga, Fiji and Samoa clustered very close to each other in the PCA, with samples from Tonga and Fiji clustering particularly close together (Fig. 3A). The beetles from all three of these countries belong to the CRB-S CoxI haplotype group (Fig 3B), and the genetic differentiation detected between them has likely occurred since they initially invaded the region. We also detected signals of admixture in the O. rhinoceros populations in Samoa, particularly Upolu, where admixture from Tonga was indicated (Fig. 4). In the structure analysis low levels of admixture from Samoa were also detected in Fiji (Fig. 4).
3.3. Incidence of OrNV infection in adult O. rhinoceros
OrNV infection was detected in adult O. rhinoceros in all countries and infection rates were extremely high (63-95% where ≥ 10 individual beetles were tested) in all haplotype groups (Table 1). We detected several OrNV infected beetles from each of the different haplotypes collected in Solomon Islands. Detection of OrNV infection in the small number of specimens collected from the remote Santa Cruz Islands demonstrates that OrNV is widely distributed in O. rhinoceros populations in Solomon Islands, but it does not provide reliable information on the prevalence of the pathogen in those outlying regions. Nonetheless, testing 96 wild caught O. rhinoceros adults collected in Guadalcanal and the Russell Islands indicated that >80% of beetles were infected with OrNV and similarly high infection rates (>90%) were recorded in O. rhinoceros collected from New Caledonia and Vanuatu (Table 1). We randomly sequenced the PCR positive fragment of virus using Sanger sequencing to make sure they do not represent false positive amplifications and these sequences have been submitted to NCBI under GenBank accession numbers MW691163-8.
Table 1.
Incidence of OrNV infection in Adult O. rhinoceros collected from the South Pacific Islands.
|
O. rhinoceros haplotype groups |
||||||||
|---|---|---|---|---|---|---|---|---|
|
O. rhinoceros origin |
CRB-S |
CRB-PNG |
CRB-G |
Overall infection rate (%) | ||||
| Country | Location | No. tested | No. OrNV +ve (%) | No. tested | No. OrNV +ve (%) | No. tested | No. OrNV +ve (%) | |
| Fiji | Viti Levu | 21 | 21 (100) | - | - | - | - | 100 |
| Vanua Levu | 17 | 15 (88) | - | - | - | - | 88 | |
| Solomon Islands | Guadalcanal, Dorma | 2 | 0 (0) | 12 | 9 (75) | 26 | 16 (64) | 63 |
| Guadalcanal, Mbalasuna | - | - | 17 | 17 (100) | 21 | 21 (100) | 100 | |
| Russell Islands | - | - | - | - | 18 | 16 (89) | 89 | |
| Gizo | - | - | 1 | 0 (0) | - | - | 0 | |
| Kolombangara | - | - | - | - | 1 | 0 (0) | 0 | |
| Santa Cruz Islands | 4 | 2 (50) | - | - | 50 | |||
| Tonga | Nuku'alofa | 20 | 19 (95) | - | - | - | - | 95 |
| Samoa | Upolu | 10 | 7 (70) | 1 | 1 (100) | - | - | 73 |
| Savai'i | 18 | 18 (100) | - | - | - | - | 100 | |
| New Caledonia | Noumea | - | - | - | - | 10 | 10 (100) | 100 |
| Vanuatu | Efate | - | - | 5 | 4 (80) | - | - | 80 |
| Papua New Guinea | New Britain, Kimbe | - | - | 15 | 11 (73) | - | - | 73 |
| Philippines | Los Baños | - | - | - | - | 9 | 7 (78) | 78 |
The dash indicates no detection of that particular haplotype in the geographical area in our sampling
4. Discussion
Understanding gene flow and genetic structure across O. rhinoceros populations can help explain active evolutionary processes occurring within the species and potentially contribute to improved pest management strategies. Mitochondrial DNA sequencing and analysis has proven extremely useful for understanding both historical and recent movement of insects, especially when divergent haplotype groups have independently invaded different islands (e.g. (Brookes et al., 2020). However, when mitochondrial data is interpreted in isolation of other genetic information, care is required because mitochondrial genetic information is only inherited maternally. When divergent mitochondrial haplotypes of a species overlap and mate, the resulting genetic admixture and geneflow between the haplotypes means that information in the mitochondrial genome is no longer a reliable marker for biological differences that may have been detected when they were in isolation (e.g. Toon et al. (2016), Brookes et al. (2020)).
By combining multiple nuclear markers with mitochondrial CoxI data and OrNV screening, this study provides greater clarity of the genetic structure of the O. rhinoceros population across the south Pacific than was previously available. It also provides a better understanding of the relationships between the different haplotype groups and OrNV. In most countries, the O. rhinoceros population is monotypic, with all individuals tested belonging to a single mitochondrial haplotype group; in Fiji and Tonga beetles were from the CRB-S haplotype group, in PNG and Vanuatu they were from the CRB-PNG group and in New Caledonia they were from the CRB-G group (Fig. 1). In Samoa, the CRB-S haplotype group dominated but an individual from CRB-PNG haplotype group was also detected. However, evidence of admixture was recorded in both islands of Samoa (Fig. 4). In Solomon Islands, all three haplotype groups were represented in the beetles analysed (Fig. 1). While the incidence of OrNV infection in samples varied, high levels of infection were confirmed in all O. rhinoceros haplotypes wherever they were found (Table 1).
In Solomon Islands, gene flow between CRB-PNG and CRB-G CoxI haplotypes is evident, as both haplotypes were present in the same geographical location and the population is a well-mixed population based on the nuclear GBS data (Fig. 4). The mitochondrial sequence data also show that the CRB-S haplotype is more closely related to the CRB-G haplotype than is the CRB-PNG haplotype, suggesting that CRB-S and CRB-G haplotypes are also likely to become admixed when they are brought into sympatry. Such a pattern has been observed in Palau where both the CRB-S and the CRB-G haplotypes are established (Reil et al. 2018, Kitalong et al. 2018). The structure analysis of Reil et al. (2018) revealed five individuals out of the 23 included from Palau (all ‘CRB-S’ CoxI haplotypes) were assigned to a somewhat different cluster to the other 18 (which were a mixture of CRB-G and CRB-S CoxI haplotypes). Both the structure and phylogenetic analyses of the GBS data indicated admixture between these two populations on Palau (Reil et al., 2018), although the authors caution that the introgression appeared to be unidirectional because they only detected CRB-S CoxI haplotypes in an historically CRB-G population. In contrast, we found extensive geneflow and admixture between the CRB-PNG and CRB-G CoxI haplotypes in Solomon Islands.
The current CoxI based haplotype diagnostics are therefore unlikely to reflect any significant biological differences between insects and they are only diagnostic for movement of beetles of a given haplotype group into a region. In places such as Solomon Islands, where multiple haplotype groups are present and there is obvious genetic admixture between them (Fig. 3), the utility of CoxI based haplotype diagnostics in even this respect is limited as beetles can only be traced back through the maternal line. In areas where the different haplotypes interbreed, these diagnostic methods cannot be used to draw meaningful conclusions about the movement of insects or their likely susceptibility to OrNV. For example, male beetles from the CRB-G CoxI haplotype that mate with female beetles from a different haplotype would produce offspring undetectable as CRB-G using CoxI haplotype diagnostics, but they would carry paternal nuclear genes, including those mediating interactions with viruses and other pathogens. Similarly, females from the CRB-G haplotype group that mate with males from another haplotype would produce offspring detectable as belonging to the CRB-G CoxI haplotype but the genetic material in their nuclear genome would be from both parents.
The genetic similarity of O. rhinoceros populations from Tonga, Samoa and Fiji probably reflects a shared invasion history in these countries. With the GBS data we do find differences between these islands, with all three being separated in the structure analysis. The admixture inferred from Samoa into Fiji, and from Tonga into Samoa (especially Upolu) could represent their recent shared ancestry, however, under that scenario one would expect the signal to be more even across the three populations. It is more likely that these signals represent some amount of migration between these islands. We found one individual from Samoa (Upolu) that clustered with Solomon Islands in the structure analysis and belonged to the CRB-G CoxI haplotype (Fig 4). Further investigation is warranted, but this individual could represent an insect that has recently arrived on Upolu from Solomon Islands. The higher admixture signal on Upolu (Samoa) makes sense as this Island is much more involved in trade compare to Savai'i. Overall, however, our population genetics analysis indicates movement between islands is relatively rare, with each island representing distinct genetic populations, despite the recent nature of the invasions.
Misinterpretation of the current pest situation, the critical genetic relationships between populations and the use of imperfect genetic markers to indicate likely tolerance to OrNV infection can confuse our primary understanding of O. rhinoceros biology and retard the development of appropriate control strategies. For example, the O. rhinoceros that have recently invaded Vanuatu and New Caledonia belong to the CRB-PNG and CRB-G CoxI haplotype groups respectively but field collected insects in both localities are positive for OrNV infection (Table 1). As in Solomon Islands, individuals responsible for these new invasions, which are the result of human mediated transport of beetles across large expanses of ocean, could be the progeny of parents from different mitochondrial haplotypes (CRB-PNG and CRB-G) but they are only identifiable based on their maternal haplotype groups. This demonstrates that genetic and phenotypic characteristics that have been associated with the CRB-G haplotype group (Marshall et al., 2017) are not restricted to these insects.
Resistance to OrNV in the CRB-G haplotype group has been proposed based on an absence of OrNV infection in wild caught specimens from this haplotype and the results from laboratory bioassays investigating the susceptibility of adult O. rhinoceros from the Guam population of this haplotype to OrNV (Marshall et. al., 2017). Contrary to our findings (Table 1), OrNV infection was not detected in O. rhinoceros belonging to the CRB-G haplotype group collected in Solomon Islands or the Philippines, but it was detected in specimens from this haplotype group collected from Palau (Marshall et al., 2017), where the CRB-G and CRB-S haplotypes occur in sympatry. Interestingly, Kitalong et al., (2018) found very high rates of OrNV infection in both haplotypes in Palau (83% in CRB-G and 92% in CRB-S), supporting our findings of a high incidence of OrNV infection in all haplotypes in the Pacific Islands and the Philippines. Rahayuwati (2020) recently reported that 90.2% and 89.5% of examined beetles in Riau and North Sumatera, Indonesia were infected with OrNV. Similarly, several studies in Malaysia have also found high incidences of virus in natural populations of beetles. PCR diagnostics have estimated the incidence of OrNV in adult O. rhinoceros collected from pheromone traps in Malaysia to be as high as 98% (Ramle et al., 2011). Estimates of OrNV incidence and infection rates in host insects are dependent on the stage of infection, sampling and collection methods, storage, and the precision of the techniques used to detect infection by the pathogen. There is no doubt that high prevalence of virus in all geographical populations of O. rhinoceros is an important finding which requires further investigation. High prevalence of persistent and asymptomatic infection has been previously reported from other members of the Nudiviridae. HzNV-2, which infects Helicoverpa zea and is closely related to OrNV, does not induce rapid mortality or immediate sterility in infected individuals that it is very prevalent and highly persistent in host populations (Burand et al., 2012).
As OrNV is highly transmissible between adult O. rhinoceros (Huger, 2005), an OrNV infected adult held in a trap for any period of time could easily transmit infection to healthy, non-infected beetles entering the trap and thereby affect estimates of the incidence of the pathogen in the wild beetle population. To confirm true infection, where virus replicates in the host, we extracted RNA from randomly selected O. rhinoceros samples and checked for the presence of virus by RT PCR (unpublished data) and we also conducted short read Illumina sequencing to provide another layer of validation (Etebari et al., 2020b). The next generation sequencing data not only confirmed the presence of virus in O. rhinoceros samples belonging to CRB-G haplotypes but also showed substantial virus replication in samples collected from Solomon Islands and Philippines (Etebari et al., 2020a; Etebari et al., 2020b). Further, low nudivirus mortality rates in infected host insects may not be related to host resistance to the pathogen but rather due to changes in the virus (Unckless, 2011).
5. Conclusion
The different haplotypes of O. rhinoceros reported in the south Pacific represent the different invasion histories of the pest in the region. The current CoxI based method is not a reliable diagnostic marker for phenotypic traits, especially in countries such as Solomon Islands where the haplotypes associated with different invasion fronts have come back into sympatry and are now mixed. Although we found OrNV infecting all CoxI haplotypes of O. rhinoceros across the Pacific, our knowledge of the host-pathogen interaction remains limited. Molecular analyses of different O. rhinoceros haplotypes in response to OrNV infection are necessary to identify potential mechanisms of resistance to OrNV. To improve biological control of O. rhinoceros, additional work is required to further improve our understanding of the population genetics of O. rhinoceros across the Pacific and to establish the evolutionary history of OrNV in the region.
Declaration of Competing Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Acknowledgements
This project was supported by the Australian Centre for International Agricultural Research funding (HORT/2016/185) and the University of Queensland (UQECR2057321). We thank Nitya N. Singh (Government of Fiji), Marie Joy Beltran (University of the Philippines Los Baños), Aurélie Chan (Department of Veterinary, Food and Rural Affairs, Government of New Caledonia) and colleagues from Papua New Guinea Oil Palm Research Association, Dami research station for assistance in collecting and providing the insect specimens for genetic analysis.
Data Accessibility
Raw genotyping by sequencing data is accessible by NCBI accession PRJNA648153. The CoxI gene sequences have been submitted to NCBI under GenBank accession numbers MN809502-MN809525. The sequence of a fragment of OrNV from different geographical regions have been submitted to NCBI under GenBank accession numbers MW691163- MW691168.
Authors' contributions
Kayvan Etebari: Conceptualization, Methodology, Investigation, Data curation, Formal analysis, Visualization, Writing- Original draft preparation, Funding acquisition. James Hereward: Methodology, Investigation, Data curation, Visualization, Writing-Reviewing and Editing. Apensia Sailo: Resources. Robert Tautua: Resources. Emeline M. Ahoafi: Resources. Helen Tsatsia: Resources. Grahame Jackson: Resources. Michael J. Furlong: Conceptualization, Investigation, Writing - Reviewing and Editing, Funding acquisition, Project administration, Supervision.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cris.2021.100015.
Contributor Information
Kayvan Etebari, Email: k.etebari@uq.edu.au.
James Hereward, Email: j.hereward@uq.edu.au.
Apenisa Sailo, Email: apenisa.sailo@govnet.gov.fj.
Emeline M. Ahoafi, Email: ahoafieme@gmail.com.
Robert Tautua, Email: robert.tautua@maf.gov.ws.
Helen Tsatsia, Email: Helen.Tsatsia@sig.gov.sb.
Grahame V Jackson, Email: grahamejackson@gmail.com.
Michael J. Furlong, Email: m.furlong@uq.edu.au.
Appendix. Supplementary materials
References
- Bedford G.O. Biological control of the rhinoceros beetle (Oryctes rhinoceros) in the South Pacific by Baculovirus. Agric. Ecosystems Environ. 1986;15:141–147. [Google Scholar]
- Bedford GO. Vol. 58. 2013. Biology and Management of Palm Dynastid Beetles: Recent Advances; pp. 353–372. (Annual Review of Entomology). Berenbaum MR, editor. Annual Review of Entomology. 58. Palo Alto: Annual Reviews. [DOI] [PubMed] [Google Scholar]
- Burand JP, Kim W, Afonso CL, Tulman ER, Kutish GF, Lu Z, et al. Analysis of the Genome of the Sexually Transmitted Insect Virus Helicoverpa zea Nudivirus 2. Viruses. 2012;4(1):28–61. doi: 10.3390/v4010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes D.R., Hereward J.P., Wilson L.J., Walter G.H. Multiple invasions of a generalist herbivore—Secondary contact between two divergent lineages of Nezara viridula Linnaeus in Australia. Evolutionary Applications. 2020;13(8):2113–2129. doi: 10.1111/eva.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caltagirone L.E. Landmark Examples in Classical Biological Control. Annual Review of Entomology. 1981;26:213–232. [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M., Gormley N., Gilbert J.A., Smith G., Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J., Hohenlohe P.A., Bassham S., Amores A., Cresko W.A. Stacks: an analysis tool set for population genomics. Molecular Ecology. 2013;22(11):3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation genetics resources. 2012;4(2):359–361. [Google Scholar]
- Elshire R.J., Glaubitz J.C., Sun Q., Poland J.A., Kawamoto K., Buckler E.S., Mitchell S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. Plos One. 2011;6(5) doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etebari K., Filipovic I., Rasic G., Devine G.J., Tsatsia H., Furlong M.J. Complete genome sequence of Oryctes rhinoceros nudivirus isolated from the coconut rhinoceros beetle in Solomon Islands. Virus research. 2020;278 doi: 10.1016/j.virusres.2020.197864. -197864. [DOI] [PubMed] [Google Scholar]
- Etebari K., Parry R., Beltran M.J.B., Furlong M.J. Transcription profile and genomic variation of Oryctes rhinoceros nudivirus (OrNV) in Coconut Rhinoceros Beetle. Journal of Virology. 2020 doi: 10.1128/JVI.01097-20. JVI.01097-01020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ero M.M., Sar S., Kawi A., Tenakanai D., Gende P., Bonneau L.J.G. Detection of the Guam biotype (CRB-G) Oryctes rhinoceros Linneaus (Coleoptera: Scarabaeidae) in Port Moresby, Papua New Guinea. Planter. 2016;92(1089):883–891. [Google Scholar]
- Ero M.M., Bonneau L.J.G. The 2018 International Congress of Invertebrate Pathology and Microbial Control and the 51st Annual Meeting of the Society for Invertebrate Pathology. 2018. Coconut Rhinoceros Beetle (CRB) control efforts in oil palm: Papua New Guinea (CRB-P) versus Solomon Islands (CRB-G) p. 49. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular ecology. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3(5):294–299. [PubMed] [Google Scholar]
- Fraenkel J., Allen M., Brock H. The resumption of palm-oil production on Guadalcanal’s northern plains. Pacific Economic Bulletin. 2010;25(1):65–75. [Google Scholar]
- Hereward J.P., Smith T.J., Brookes D.R., Gloag R., Walter G.H. 2020. Tests of hybridisation in Tetragonula stingless bees using multiple genetic markers. bioRxiv. 2020.2003.2008.982546. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huger A.M. The Oryctes virus: Its detection, identification, and implementation in biological control of the coconut palm rhinoceros beetle, Oryctes rhinoceros (Coleoptera: Scarabaeidae) Journal of Invertebrate Pathology. 2005;89(1):78–84. doi: 10.1016/j.jip.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Kitalong C., Ramarui J.O., Ngiramengior J.Skey, B. Masang, N. Watanabe, S. Melzer, M. Nakai, amd J Miles M. The 2018 International Congress of Invertebrate Pathology and Microbial Control and the 51st Annual Meeting of the Society for Invertebrate Pathology. 2018. CRB damage and resistance assessment in the Palau Archipelago; p. 48. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources. 2015;15(5):1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J.W., Bryant D. POPART: full-feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6(9):1110–1116. [Google Scholar]
- Linck E., Battey C.J. Minor allele frequency thresholds strongly affect population structure inference with genomic data sets. Molecular Ecology Resources. 2019;19(3):639–647. doi: 10.1111/1755-0998.12995. [DOI] [PubMed] [Google Scholar]
- Marschal K. Introduction of a new virus disease of the Coconut Rhinoceros Beetle in Western Samoa. Nature. 1970;225(5229):288–289. [Google Scholar]
- Marshall S.D.G., Moore A., Vaqalo M., Noble A., Jackson T.A. A new haplotype of the coconut rhinoceros beetle, Oryctes rhinoceros, has escaped biological control by Oryctes rhinoceros nudivirus and is invading Pacific Islands. Journal of Invertebrate Pathology. 2017;149:127–134. doi: 10.1016/j.jip.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Peterson B.K., Weber J.N., Kay E.H., Fisher H.S., Hoekstra H.E. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. Plos One. 2012;7(5) doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland J.A., Brown P.J., Sorrells M.E., Jannink J.L. Development of High-Density Genetic Maps for Barley and Wheat Using a Novel Two-Enzyme Genotyping-by-Sequencing Approach. Plos One. 2012;7(2) doi: 10.1371/journal.pone.0032253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahayuwati S, Kusumah YM, Prawirosukarto S, Dadang Santoso T. The status of Oryctes rhinoceros Nudivirus (OrNV) infection in Oryctes rhinoceros (Coleoptera: Scarabaeidae) in Indonesia. Journal of Oil Palm Research. 2020;32(4):582–589. [Google Scholar]
- Ramle M., Kamarudin N., Ghani I.A., Wahid M.B., Jackson T.A., Tey C.C., Ahdly M. Molecular approaches in the assessment of Oryctes rhinoceros virus for the control of rhinoceros beetle in oil palm plantations. Journal of Oil Palm Research. 2011;23:1096–1109. [Google Scholar]
- Reil J.B., Doorenweerd C., San Jose M., Sim S.B., Geib S.M., Rubinoff D. Transpacific coalescent pathways of coconut rhinoceros beetle biotypes: Resistance to biological control catalyses resurgence of an old pest. Molecular Ecology. 2018;27(22):4459–4474. doi: 10.1111/mec.14879. [DOI] [PubMed] [Google Scholar]
- Reil J.B., San J., Rubinoff D. Low variation in nuclear and mitochondrial DNA inhibits resolution of invasion pathways across the Pacific for the Coconut Rhinoceros Beetle (Scarabeidae: Oryctes rhinoceros) Proceedings of the Hawaiian Entomological Society. 2016;48:57–69. [Google Scholar]
- Richards N.K., Glare T.R., Aloali'i I., Jackson T.A. Primers for the detection of Oryctes virus from Scarabaeidae (Coleoptera) Molecular Ecology. 1999;8(9):1552–1553. doi: 10.1046/j.1365-294x.1999.07072.x. [DOI] [PubMed] [Google Scholar]
- Ridley A.W., Hereward J.P., Daglish G.J., Raghu S., McCulloch G.A., Walter G.H. Flight of Rhyzopertha dominica (Coleoptera: Bostrichidae)-a Spatio-Temporal Analysis With Pheromone Trapping and Population Genetics. Journal of Economic Entomology. 2016;109(6):2561–2571. doi: 10.1093/jee/tow226. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Toon A., Daglish G.J., Ridley A.W., Emery R.N., Holloway J.C., Walter G.H. Random Mating Between Two Widely Divergent Mitochondrial Lineages of Cryptolestes ferrugineus (Coleoptera: Laemophloeidae): A Test of Species Limits in a Phosphine-Resistant Stored Product Pest. Journal of Economic Entomology. 2016;109(5):2221–2228. doi: 10.1093/jee/tow178. [DOI] [PubMed] [Google Scholar]
- Tsatsia F., Tsatsia H., Wratten H., Macfarlane B. Solomon Islands. 51st Annual meeting of the society for invertebrate pathology. 2018. The status of Coconut Rhinoceros Beetle, Oryctes rhinoceros (L) Scarabaeidae: Dynastinae; p. 49. [Google Scholar]
- Unckless R.L. A DNA Virus of Drosophila. Plos One. 2011;6(10) doi: 10.1371/journal.pone.0026564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny B. Occurrence of the baculovirus disease of the coconut palm rhinoceros beetle in the Philippines and in Indonesia. FAO Plant Prot. Bull. 1977;25:73–77. [Google Scholar]
- Zelazny B. Oryctes rhinoceros populations and behavior influenced by a baculovirus. Journal of Invertebrate Pathology. 1977;29(2):210–215. [Google Scholar]
- Zelazny B., Alfiler A.R. Ecology of baculovirus-infected and healthy adults of Oryctes rhinoceros (Coleoptera: Scarabaeidae) on coconut palms in the Philippines. Ecological Entomology. 1991;16(2):253–259. [Google Scholar]
- Zelazny B., Lolong A., Crawford A.M. Introduction and field comparison of baculovirus strains against Oryctes rhinoceros (Coleoptera: Scarabaeidae) in the Maldives. Environmental Entomology. 1990;19(4):1115–1121. [Google Scholar]
- Zelazny B., Lolong A., Pattang B. Oryctes rhinoceros (Coleoptera: Scarabaeidae) populations suppressed by a baculovirus. Journal of Invertebrate Pathology. 1992;59(1):61–68. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw genotyping by sequencing data is accessible by NCBI accession PRJNA648153. The CoxI gene sequences have been submitted to NCBI under GenBank accession numbers MN809502-MN809525. The sequence of a fragment of OrNV from different geographical regions have been submitted to NCBI under GenBank accession numbers MW691163- MW691168.