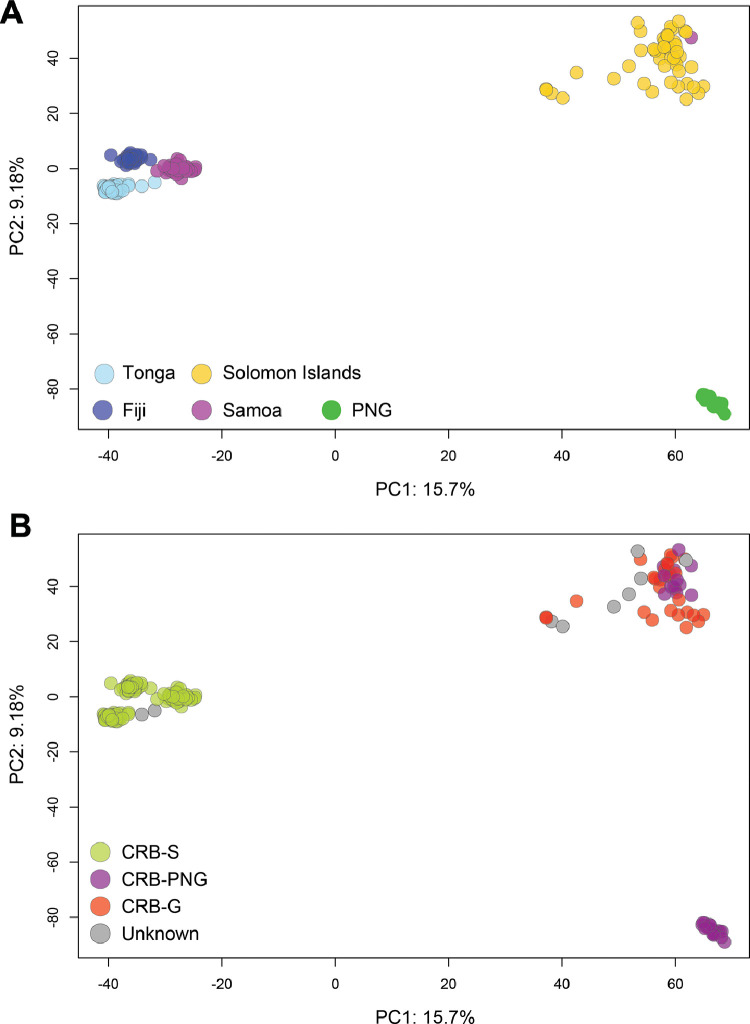
Fig. 3.
Principal component analysis (PCA) plots of the GBS data coloured by country of origin (A), and by CoxI haplotype groups (B). We produced a variant call file (vcf) using the populations command in stacks and applying a heterozygosity threshold of 0.65 with multiple snps per locus. We plotted a PCA using the adegenet package in R (Jombart, 2008), based on 6,561 snps. We couldn't confirm the CoxI haplotypes of some individuals due to low quality of reads from sanger sequencing and marked them as “unknown” in PCA plot.