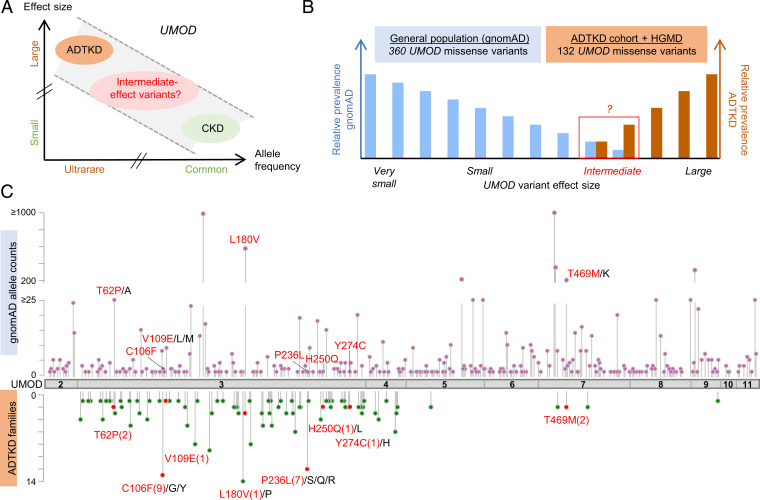
Fig. 1.
Identification of intermediate-effect UMOD variants. (A) Genetic architecture of UMOD-associated diseases showing the continuum of CKD risk associated with ultrarare (AF < 10−4) high-effect to common (AF > 0.05) low-effect UMOD variants. Adapted from ref. 17. (B) Schematic representation of the study design; 360 distinct missense UMOD variants are reported in the gnomAD with an individual AF of 2.0 × 10−2 to 3.6 × 10−6, and 132 distinct missense UMOD variants reported in ADTKD-UMOD have been extracted from the International ADTKD Cohort (726 patients from 585 families) and from the HGMD. The theoretical maximum credible AF of high-effect UMOD variants (pathogenic) in the overall population considering the disease prevalence, allelic and genetic heterogeneity, and a penetrance of 100% is estimated to be 1 × 10−7 (46) (SI Appendix), excluding those (in theory) from gnomAD. Plotted here are the expected relative prevalences of very small– to large–effect size UMOD variants in gnomAD and reported in ADTKD-UMOD patients. Conferring nonfully penetrant ADTKD or reduced disease expressivity, intermediate-effect UMOD variants might be found overlapping at the lower prevalence spectrum in both gnomAD and reported variants in ADTKD-UMOD. (C) The UMOD protein (NM_003361) is in gray with exon boundaries marked by dotted lines. UMOD missense variants reported in the gnomAD consortium are plotted on top of the protein with respect to the amino acid position. The y axis represents gnomAD allele counts. UMOD missense variants reported in the International ADTKD Cohort and in HGMD are plotted below the protein. The y axis represents the number of reported families. Variants that are reported both in gnomAD and in at least one ADTKD family are marked in red (with family numbers in parentheses). Protein and variant visualization used ProteinPaint (47) and cBioPortal MutationMapper (48, 49).