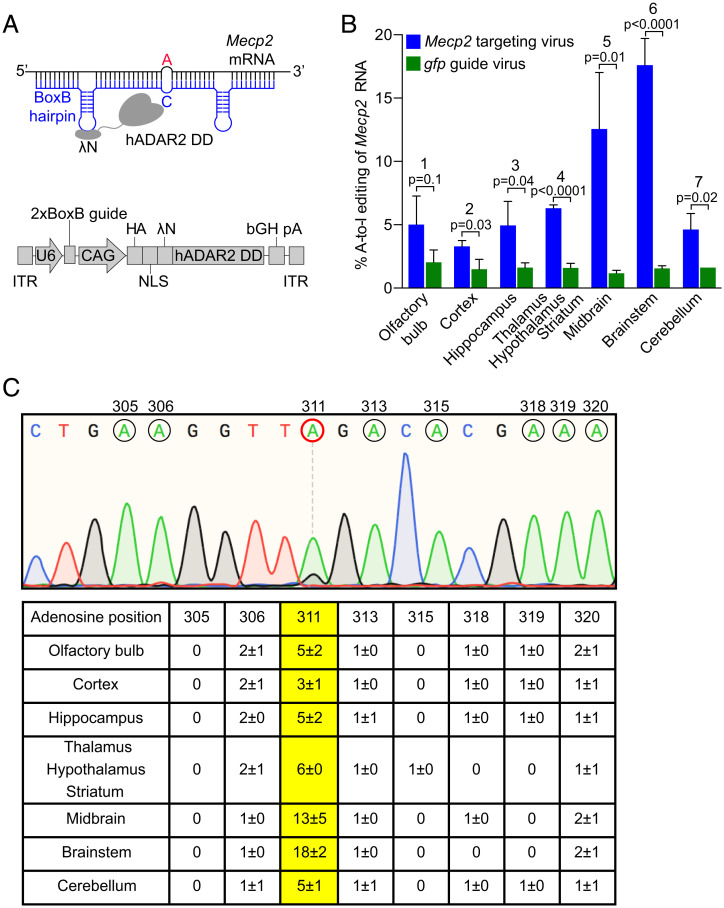
Fig. 2.
Essential components of the targeted RNA editing strategy and resulting on-target editing frequencies at Mecp2G311A. (A, Upper) The hybrid Editasewt consists of the bacteriophage λN RNA binding peptide, fused to the native human ADAR2 deaminase domain (hADAR2 DD). A guide RNA (blue) contains 46 nucleotides complementary to Mecp2 mRNA (black) and two copies of the BoxB hairpin from bacteriophage λ, recognized by the λN RNA binding peptide in Editase. As a control for the specificity of editing, a 2xBoxB guide containing sequences complementary to gfp RNA (gfp guide) was substituted for the Mecp2-targeting guide. To increase editing efficiency, opposite of the target A (red), a C is introduced into the guide RNA. (Lower) Schematic of AAV encoded RNA editing components. U6, human U6 promoter; CAG, CMV enhancer, chicken β-actin promoter. Two copies of an HA-epitope tag, and three copies of an NLS, are added to the amino terminus of the Editasewt enzyme. (B) Quantification of on-target RNA-editing percentages based on Sanger sequencing (mean ± SD, n = 3 mice per condition). Bracketed P values above each region represent editing percentages compared between the two viral conditions (blue and green). Statistics are by unpaired two-tailed t test. Numbers above each brain region refer just to the mice injected with Mecp2-targeting virus (blue), and indicate pairwise comparisons of editing percentages across the brain regions. (1, 2; ns), (1, 3; ns), (1, 4; ns), (1, 5; **), (1, 6; ***), (1, 7; ns), (2, 3; ns), (2, 4; ns), (2, 5; ***), (2, 6; ****), (3, 4; ns), (3, 5; ***), (3, 6; ****), (3, 7; ns), (4, 5; *), (4,6; ****), (4, 7; ns), (5, 6; *), (5, 7; **), (6, 7; ****). ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by repeated-measures one-way ANOVA and Tukey’s multiple comparisons test. (C, Upper) Representative Sanger sequencing chromatogram of cDNA from brainstem of a mouse injected with the Mecp2-targeting virus. Adenosines within the guide region of Mecp2 are indicated by black circles with the base number listed above. Target A is encircled in red. (Lower) Quantification of RNA editing percentages (mean ± SD, n = 3 mice per condition) for all adenosines within the guide region of Mecp2 across each brain region. Editing frequencies of the target A are in yellow.