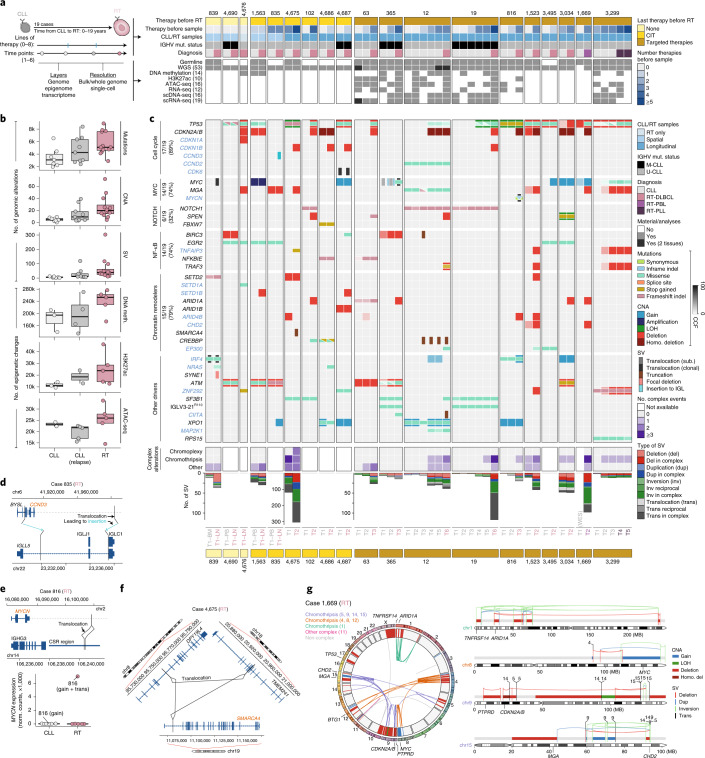
Fig. 1. The genomic landscape of RT.
a, Summary of the study. mut., mutation. b, Increase in genomic alterations and epigenetic changes compared to healthy naive and memory B cells over the disease course. Center line indicates median; box limits indicate upper and lower quartiles; whiskers indicate 1.5 × interquartile range; and points indicate individual samples. c, Driver alterations of CLL and RT. New drivers in RT are labeled in blue. Each column represents a sample and genes are represented in rows. The transparency of the color of mutations and CNAs indicates the cancer cell fraction (CCF). The number of tumors harboring an alteration at the time of transformation is indicated for each biological group of drivers (left). Complex structural alterations are shown below, together with the total number of SVs. LOH, loss of heterozygosity. d, Schema of the CCND3 insertion next to the constant region IGLC1 in the RT sample of patient 835. e, Reciprocal translocation between MYCN and class-switch recombination (CSR) region of IGHG3 in the RT sample of patient 816 (top). MYCN expression based on bulk RNA-seq (bottom). f, Chromoplexy disrupting SMARCA4 in the RT sample of patient 4,675. g, The circos plot (left) displays the SVs (links) and CNAs (inner circle) found in the RT sample of patient 1,669. CNAs are colored by type and SVs are colored according to their occurrence within specific complex events. Target driver genes are annotated. Chromosome-specific plots (right) illustrate selected complex rearrangements affecting one or multiple driver genes with CNAs and SVs colored by type.