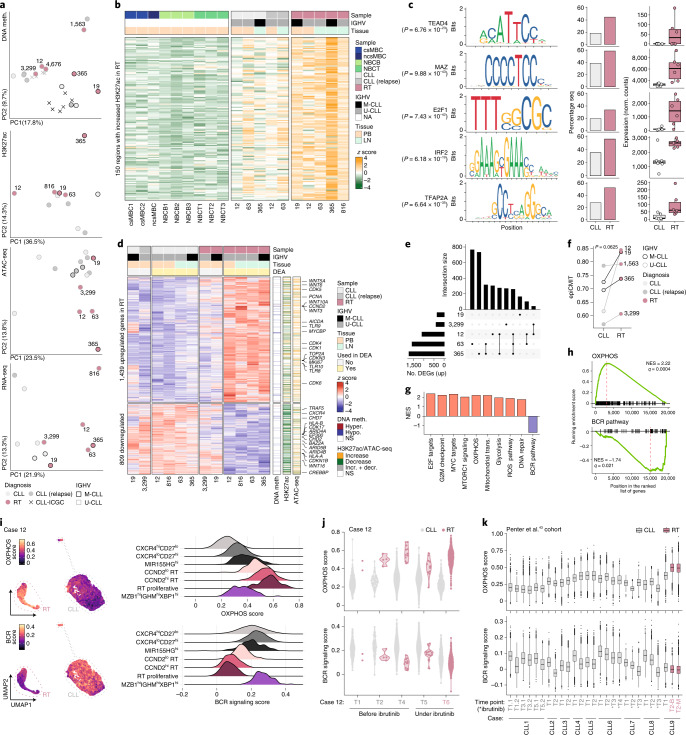
Fig. 4. Proliferation, OXPHOS and BCR pathways dominate the epigenome and transcriptome of RT.
a, PCA of the bulk epigenetic and transcriptomic layers analyzed. b, Heat map showing 150 regions with increased H3K27ac levels in RT. c, TF enriched within the ATAC peaks identified in the regions of increase H3K27ac in RT. The motif, percentage of RT-specific active regions and regions with increased H3K27ac in CLL that contained the motif and TF expression (bulk RNA-seq) in CLL and RT are shown. Center line indicates median; box limits indicate upper and lower quartiles; whiskers indicate 1.5 × interquartile range; points indicate individual samples. P values were derived using a one-tailed Wilcoxon rank-sum test. d, Heat map showing the DEGs between CLL and RT identified by bulk RNA-seq. Samples used in the differential expression analysis (DEA) are indicated. The overlap of DEGs with DNA methylation changes, H3K27ac and ATAC peaks is shown on the right. Selected genes are annotated. e, Intersection of upregulated genes in RT compared to CLL in scRNA-seq analyses. f, epiCMIT evolution from CLL to RT. P values were derived by paired Wilcoxon signed-rank test. g, Summary of the main gene sets modulated in RT based on bulk RNA-seq. NES, normalized enrichment score; ROS, reactive oxygen species. h, Gene set enrichment plot for OXPHOS and BCR signaling (bulk RNA-seq). i, OXPHOS and BCR signaling scores depicted at single-cell level for case 12 (all time points together). RT and CLL cells are highlighted (left). Ridge plots show the OXPHOS and BCR score across clusters (right). j, OXPHOS and BCR signaling scores of CLL and RT cells of patient 12 across time points by scRNA-seq. k, Distribution of OXPHOS and BCR signaling scores at a single-cell level across different time points of nine cases included in the study of Penter et al.43. Center line indicates median; box limits indicate upper and lower quartiles; whiskers indicate 1.5 × interquartile range; points indicate outliers. B, peripheral blood; M, bone marrow. *Sample collected under treatment with ibrutinib.