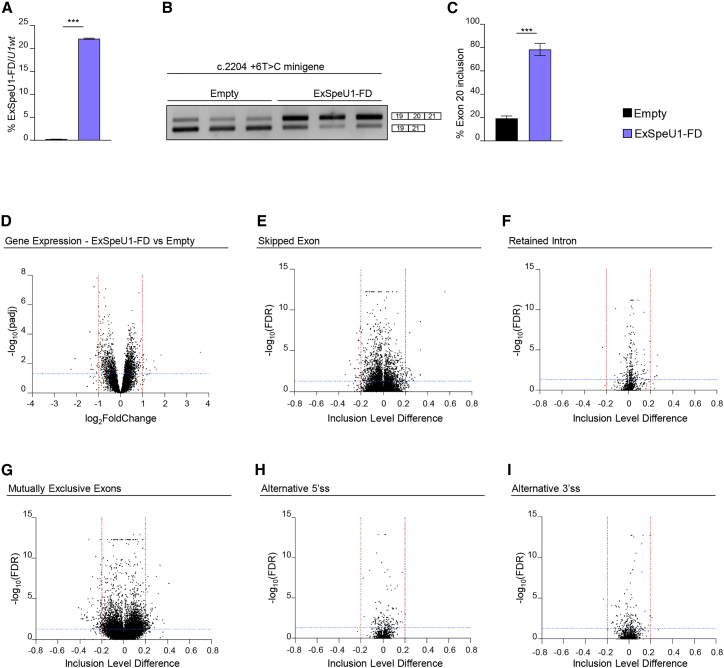
Figure 5.
Lack of global effects of ExSpe-U1-FD on the human transcriptome
(A) ExSpe-U1-FD expression levels in HEK 293 Flp-In T-REx stable control cells (empty) (n = 6) and stable clone cell line expressing ExSpe-U1-FD (n = 6). Data are expressed as percentage of ExSpe-U1-FD relative to endogenous U1.
(B) Transfection experiment of ELP1 mutant (c.2204+6T>C) minigene in ExSpe-U1-FD stable clones and controls. The ELP1 exon 20 inclusion and exclusion bands are indicated.
(C) Quantification of the percentage of exon 20 inclusion in the ELP1 mutant (c.2204+6T>C) minigene transfection experiment shown in (B).
(D) Volcano plot showing global gene expression changes between controls (empty, n = 6) and ExSpe-U1-FD stable clones (n = 6). Horizontal blue line (padj <0.05) and vertical red lines (log2FoldChange ≤−1 or ≥1) indicate cut-off values and determine significant down- and up-regulated events, respectively.
(E–I) Volcano plots showing global alternative splicing changes in (E) skipped exon (SE), (F) retained intron (RI), (G) mutually exclusive exons (MXE), (H) alternative 5′ splice site (A5SS), and (I) alternative 3′ splice site (A3SS) categories between empty clones (n = 6) and ExSpe-U1-FD stable clones (n = 6). Horizontal blue line (FDR ≤ 0.05) and vertical red lines (inclusion level difference ≤−0.2 or ≥0.2) indicate cut-off values and determine significant events in each category. In (A) and (C), data are expressed as mean ± SD and statistical analysis was performed using Student’s t test (∗∗∗p < 0.001).