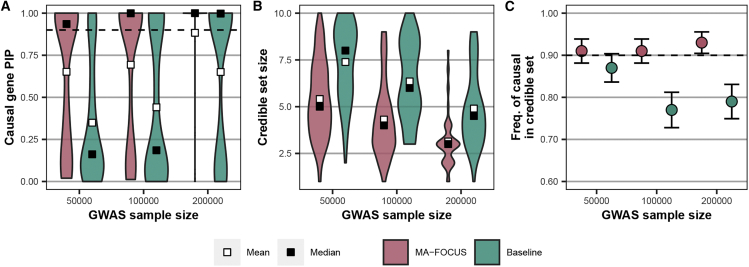
Figure 2.
MA-FOCUS outperforms the baseline approach in all three metrics as GWAS sample sizes vary when eQTLs are independent across ancestries
(A–C) Posterior inclusion probabilities (PIPs) for 100 simulated causal genes (A), the distribution of 90% credible set sizes for 100 simulated gene regions (B), and the sensitivity (C) from MA-FOCUS, and baseline approach, varying genome-wide association study (GWAS) sample sizes across multi-ancestry ancestries. See the material and methods for default parameters. The black dashed lines indicate 90%. Error bars are constructed using a 95% confidence interval.