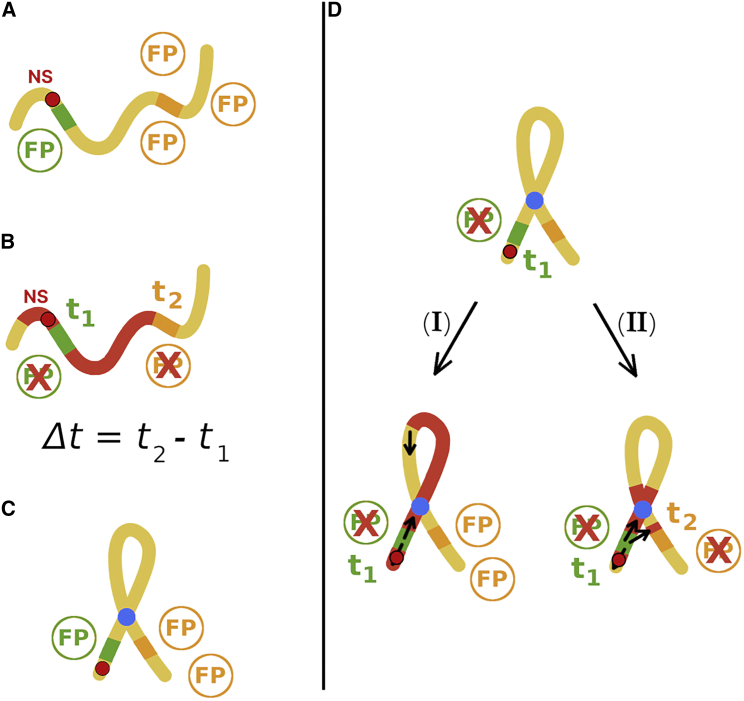
Figure 7.
Scheme for the proposed experiment. (A) The gene for the orange fluorescent protein (OFP, in orange) is expressed if chromatin is unmarked (yellow). The repression of the green fluorescent protein gene (GFP) acts as a proxy for the binding of epigenetic spreading initiators to the NS, shown in a red circle with a black border. Because the nucleation event has not occurred, the GFPs and OFPs are produced. The presence of fluorescent proteins (FPs) could be detected by measuring fluorescence intensity at a given wavelength. (B) The binding of epigenetic spreading initiators to the NS represses the GFP expression at time t1, which could be measured. Subsequently, epigenetic markers (red) spread until the OFP gene is inactivated, resulting in the disappearance of orange fluorescence at t2, which is measurable. During spreading, chromatin can form loops but need not have a permanent structure. The time difference, Δt = t2−t1, could be measured for many single cells to obtain the distribution P(Δt). This represents the control experiment; a similar experimental setup has been employed in (44). (C) A permanent loop (blue) in chromatin structure may be constructed so that the spatial proximity of the NS and OFP gene is decreased for a given genomic NS-FP distance. (D) Using the FPs as reporters for spreading, only cells that lack green fluorescence are taken into account, as it marks the successful initiation of epigenetic spreading from the NS. If the heterochromatin spreading from the NS is strictly 1D, then P(Δt) should not differ from the result in the control experiment. If 3D spreading is relevant, P(Δt) would differ compared with the control experiment. The same conclusions should be achieved by placing the OFP gene at different genomic distances from the NS, for a given loop length. To see this figure in color, go online.