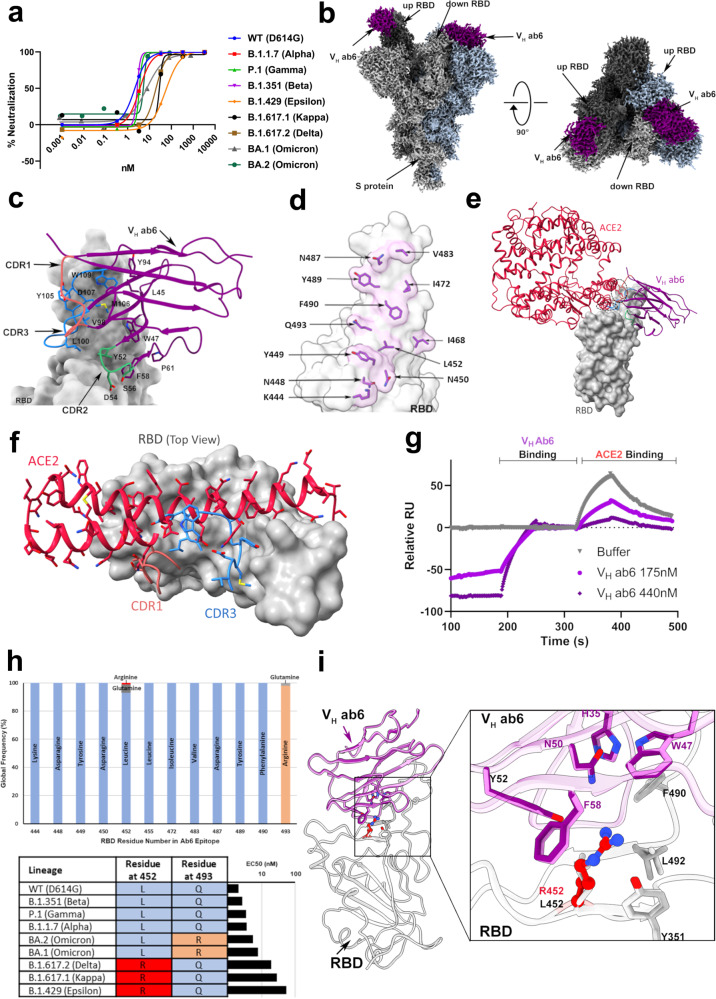
Fig. 1. Ab6 broadly neutralizes SARS-CoV-2 variants via a largely conserved molecular epitope.
a Pseudovirus neutralization of SARS-CoV-2 variants by VH ab6, performed in at least technical triplicate (n = 3), the mean is plotted. b 2.4 Å global cryo-EM density map of VH ab6 bound to wild-type S protein. Density corresponding to S protein protomers and ab6 are shown in grayscale and blue, respectively. c ab6 contact zones. The RBD and ab6 are shown as a gray surface and colorized cartoon, respectively. The ab6 scaffold is colored purple and complementarity determining regions (CDRs) of ab6 are colored as follows: CDR1—red; CDR2—green; CDR3—blue. d Footprint of ab6. The sidechains of footprint residues are shown in purple. e Overlap of ab6 and ACE2 binding footprints. The local refined model of the ab6-RBD interface was superposed with the crystal structure of the ACE2-RBD complex (PDB: 6M0J). ACE2 is shown in red while VH ab6 is shown as in c. The RBD is depicted as a gray surface. Models were aligned using the RBD. Ovals highlight steric clashing between ACE2 and VH ab6. f Detailed view of clashes made by CDR3 and CDR1 of VH ab6 with the N terminal helices of ACE2. g SPR-based spike protein competition assay between ACE2 and VH ab6. Spike protein was loaded onto an SPR chip surface before buffer or indicated concentrations of VH ab6 were injected, followed by injection of ACE2-FC. Relative response units (RUs) are plotted on the Y axis. h (Top) Global frequency of residue identity within the ab6 footprint in GISAID deposited sequences as of May 1st, 2022. (Bottom) Residue identity at positions 452 and 493 within SARS-CoV-2 variants and VH ab6 half-maximal effective concentrations (EC50) from pseudoviral neutralization assays. i Focused view superpositions of the cryo-EM-derived atomic model of the Epsilon (B.1.429) and wild-type (D614G) S proteins bound to VH ab6. Epsilon and wild-type RBDs are colored light and dark gray respectively, while purple and pink models refer to ab6-WT and ab6-Epsilon, respectively. The R452 mutation is highlighted in red. Source data are provided as a Source data file.