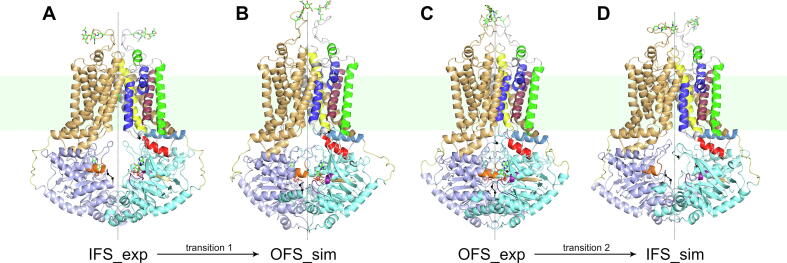
Fig. 4.
Conformational transitions during the ketMD simulations. (A) The experimental structure (PDB 6HCO) with the modelled missing loops and the added two ATP-Mg2+ that was used (after equilibration) as starting structure for the ketMD simulation of transition 1, (B) the final simulated conformation of transition 1, (C) the experimental structure (PDB 6HBU) with the modelled missing loops that was used (after equilibration) as starting structure for the ketMD simulations of transition 2 (either with bound ATP-Mg2+, ADP, or no bound nucleotides), (D) the final simulated conformation of transition 1 (in the absence of bound nucleotides). The ATPs are in licorice, the Mg2+ ions in sphere representation. The rotational symmetry axis of the homodimer is indicated by dashed lines.