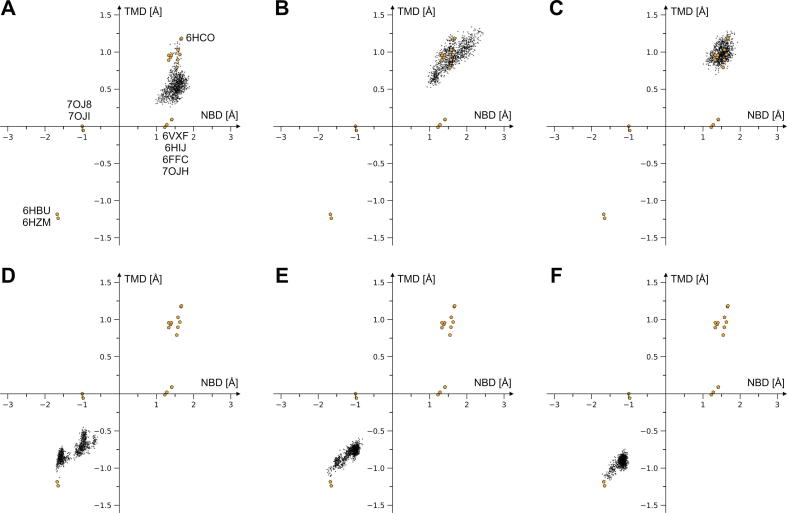
Fig. 8.
The classical MD generated conformations projected in the subspace of the NBDs’ and TMDs’ difference vectors. The difference vector, after overlapping the IFS (PDB 6HCO) and the OFS (PDB 6HBU) experimental structures, points for each Cα atom from its 3D coordinates in the OFS to its position in the IFS structure. The NBDs and TMDs difference vectors were used for the projection of the conformational differences from the mean experimental structure (of PDB 6HCO and 6HBU) in the case of (A) the apo IFS, (B) the substrate-bound IFS, and (C) the substrate- and ATP-Mg2+-bound IFS transporter, (D) the ATP-Mg2+-bound OFS, (E) the ADP-bound OFS, and (F) the nucleotide-free OFS ABCG2. Available experimental structures are marked with orange pentagons. The following experimental structures, which fall in the IFS region, are shown but not labelled: PDB 5NJ3, 6ETI, 6FEQ, 6VXI, 6VXJ, 7NEQ, 7NEZ, 7NFD. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)