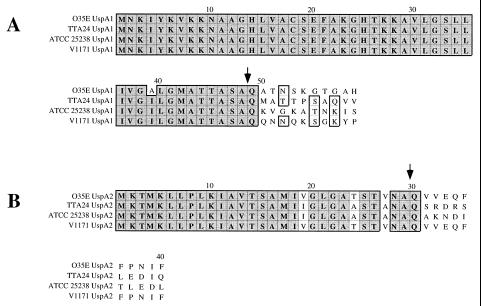
FIG. 1.
Comparison of the deduced amino acid sequence of the N-terminal regions of the UspA1 and UspA2 proteins from four M. catarrhalis strains. The cleavage sites determined for the UspA1 and UspA2 proteins from strain O35E are indicated with arrows. The signal peptide for UspA1 was 48 residues in length whereas that for UspA2 contained 29 residues. The amino acid sequence comparison was made by using the Clustal-W Alignment program in MacVector, version 6. Dark shading indicates residues that are identical. Light shading indicates residues that are similar.