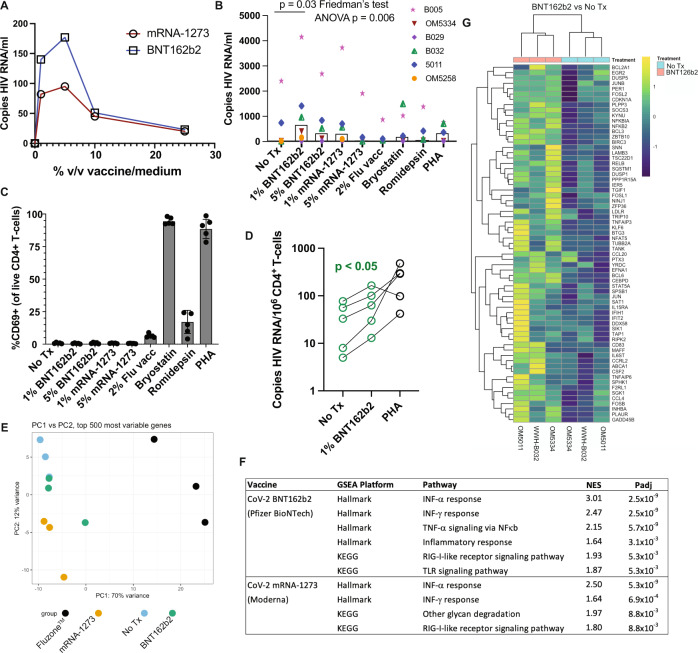
Fig. 1. The BNT162b2 mRNA vaccine stimulates the RIG-I/TLR – TNF-α – NFκb axis and activates HIV transcription ex vivo.
A qPCR measurements of HIV RNA in supernatants, 48 hours following ex vivo treatment of PBMCs from an ART-treated participant with the indicated concentrations of BNT126b2 (Pfizer BioNTech) or mRNA-1273 (Moderna) mRNA vaccines. B Extension of results from A to n = 6 ART-treated participants, adding treatments with 2% volume/volume Fluzone™ influenza vaccine, 25 nM bryostatin-1, 40 nM romidepsin, or 2 μg/ml phytohemagglutinin-L (PHA). P values were calculated by Friedman test with Dunn’s multiple comparison test (two-tailed). C Flow cytometry data from the same samples n = 5 donors harvested for (B) Shown are % CD69+ (activated) following gating on viable CD4+ T-cells. Data are presented as mean values +/− SD. D Cell-associated HIV RNA measures from the same samples as (B) P value of 0.0487 was calculated by a two-tailed paired t test between No Tx and 1% BNT162b2. E–G Bulk mRNA-seq data was generated using a subset of the samples plotted in (WWH-B032, OM5011, and OM5334). B, E Principal component analysis (PCA). The results show that transcriptional profiles of BNT126b2- and mRNA-1273-treated cells are more similar to ‘No treatment’ and to each other than Fluzone™-treated cells. F Gene set enrichment analyses showing pathways activated following mRNA vaccine treatments. Benjamini–Hochberg corrected P-values were calculated with the fgsea packge by a two-sided Weighted Kolmogorov–Smirnov (WGS) test. G Heatmap of 67 genes in the leading edge for the TNFA_SIGNALING_VIA-NFKB pathway, comparing BNT126b2 to No treatment. Source data are provided as a Source Data file.