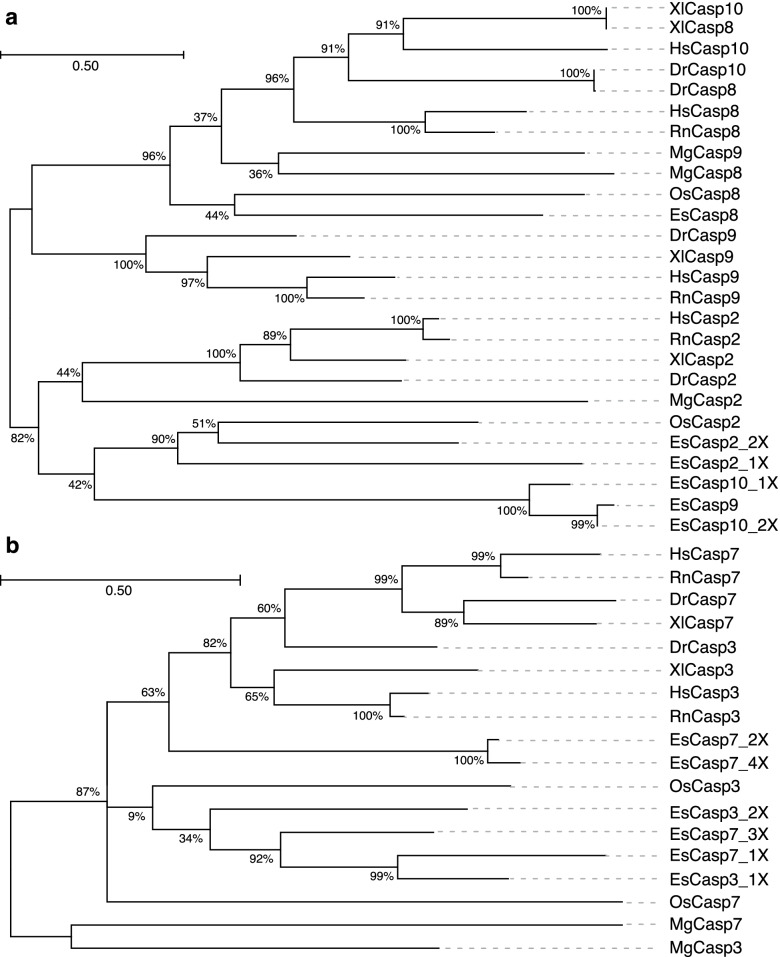
Fig. 6.
Maximum likelihood phylogenetic analysis of the caspase enzymes in E. scolopes, H. sapiens, R. norvegicus, D. rerio, X. laevis, M. galloprovincialis, and O. sinensis. a Unrooted phylogenetic tree for initiator caspase-2, -8, -9, and -10. b Unrooted phylogenetic tree for executioner caspase-3 and -7. Both trees were generated assuming the WAG model of amino acid substitution, with 1000 bootstrap iterations, in MEGA X. Branch support values are expressed as percentages. Sequences are labeled with the first letter of the corresponding genus and species (e.g., Euprymna scolopes, Es)