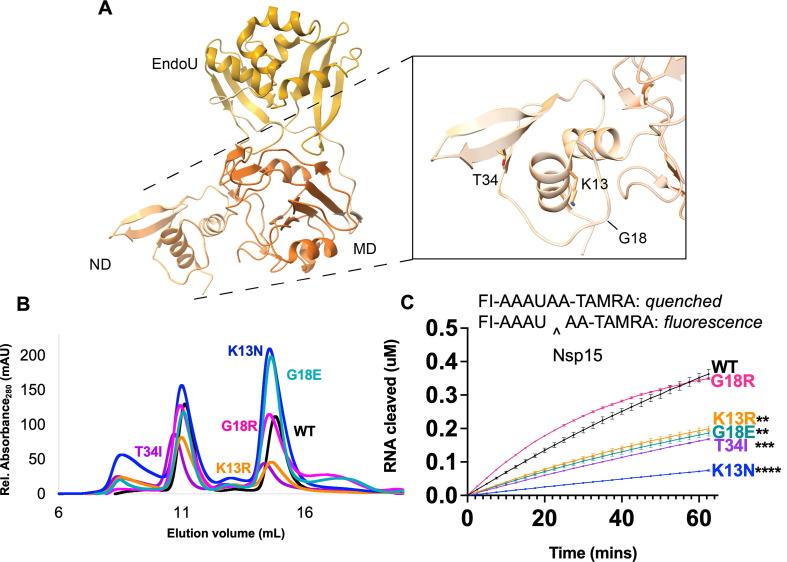
Figure 3.
Characterization of Nsp15 N terminal domain variants. (A) Displayed on the structure are the residues K13, G18, and T34. (B) S200 elution profiles of NTD variants. Hexameric Nsp15 elutes at 11 mL, monomeric Nsp15 at 15 mL. (C) An embedded visual of the Fluorescence resonance energy transfer cleavage assay to accompany the FRET time course data for Nsp15 N terminal domain mutants. Nsp15 WT and variants (2.5 nM) were incubated with RNA (0.8 μM) at room temperature and fluorescence was monitored every 2.5 min for an hour. The average of a representative technical triplicate is plotted with standard deviation error bars. At least two biological replicates were performed for each mutant. Each mutant is represented by a different color: WT Nsp15 (black), Nsp15 K13N (blue), Nsp15 K13R (orange), Nsp15 G18E (teal), Nsp15 G18R (pink), and Nsp15 T34I (purple). **p < 0.01, ***p < 0.001, ****p < 0.0001.