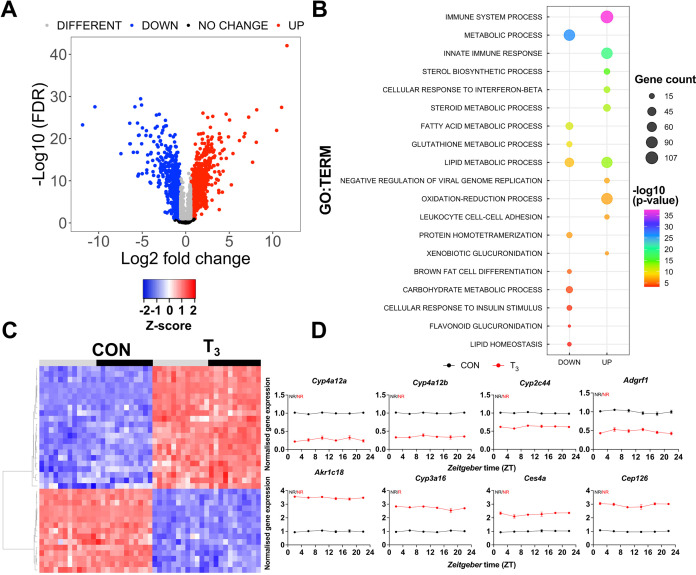
Figure 2. Identification of daytime-independent differentially expressed genes (DEGs) in the liver of triiodothyronine (T3) mice.
(A) Global (all Zeitgeber times [ZTs] included) evaluation of liver transcriptomes revealed 2336 DEGs of which 1391 and 945 were considered as up- or downregulated, respectively, using a false discovery rate (FDR) < 0.1. Genes with an FDR <0.1 were classified as different irrespectively of fold change values. (B) Top 10 list of biological processes from gene set enrichment analysis (GSEA) of up- and downregulated DEGs is represented. Additional processes can be found in Supplementary file 3. (C) Heatmap of liver DEGs showing significant T3-dependent regulation across all timepoints. Light and dark phases are shown as gray and black, respectively. (D) Diurnal expression profiles of most robustly regulated DEGs. Gene expression of all groups was normalized by CON mesor. Additional information is described in Supplementary file 4. None of these genes showed rhythmic regulation across the day (NR). n = 4 samples per group and timepoint, except for the T3 group at ZT 22 (n = 3).