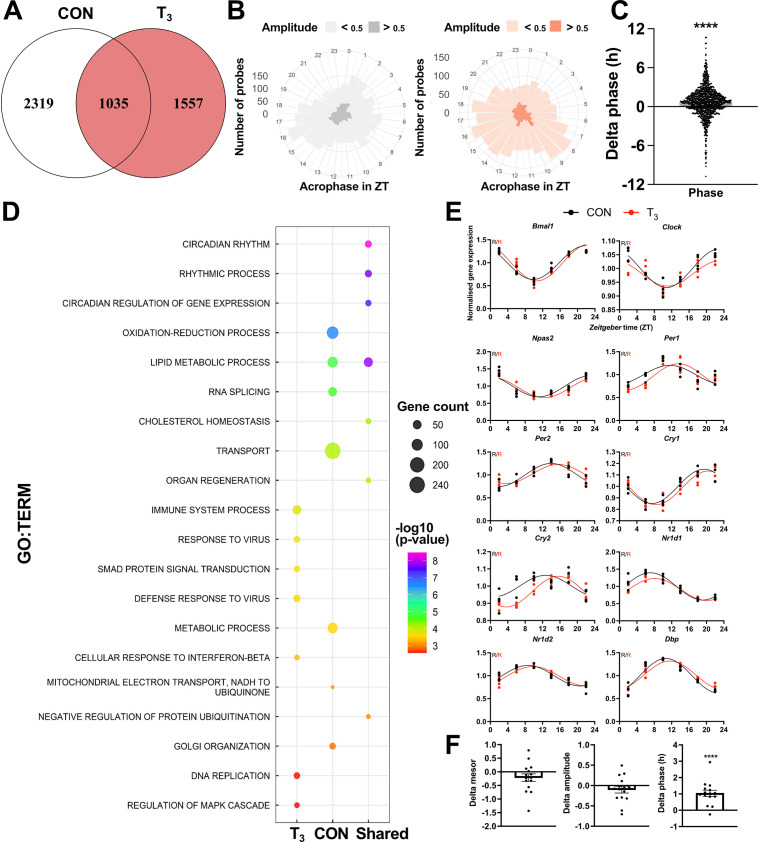
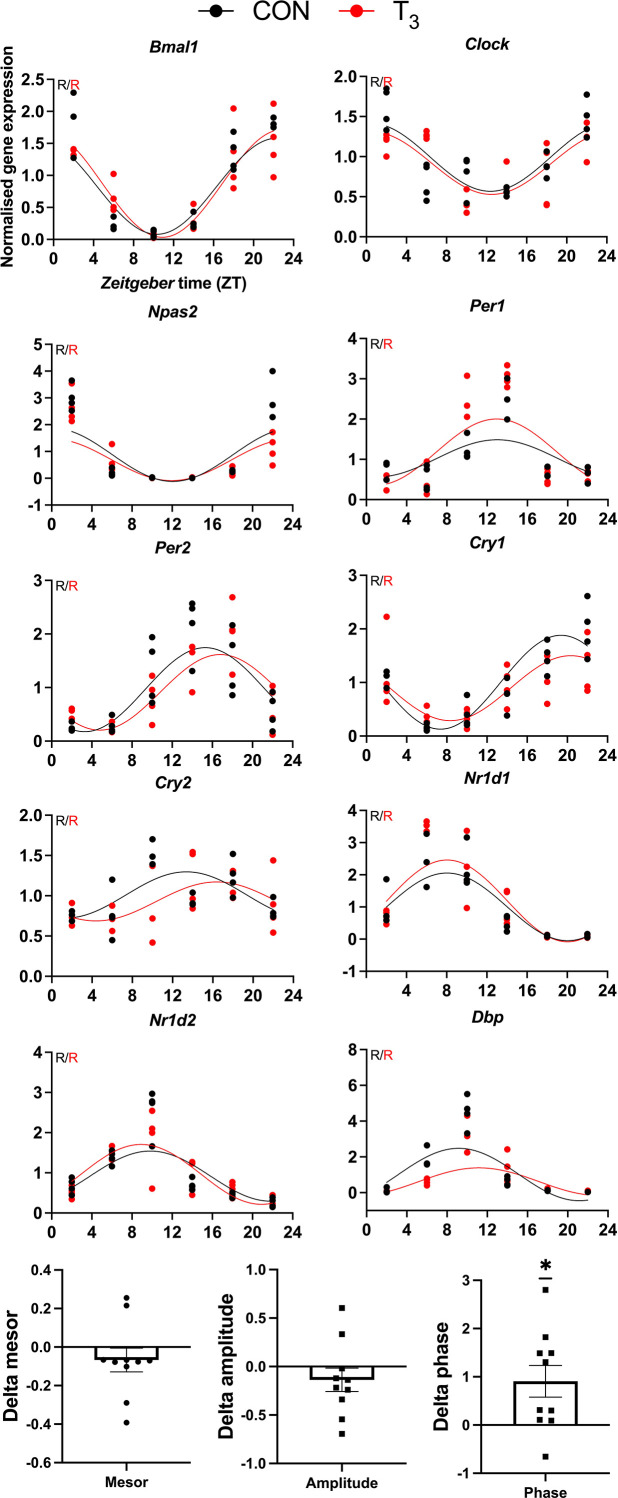
Figure 3. Diurnal evaluation of liver transcriptome of triiodothyronine (T3) mice.
(A) Rhythmic probes were identified using the JTK_CYCLE algorithm (Supplementary file 5). Venn diagram represents the distribution of rhythmic probes for each group. (B) Rose plot of all rhythmic genes from control (CON) (gray) and T3 (red) are represented by the acrophase and amplitude. Phase estimation was obtained from CircaSingle algorithm. (C) Phase difference between shared rhythmic genes. Each dot represents a single gene. One-sample t-test against zero value was performed and a significant interaction (mean 0.7781, p<0.001) was found. (D) Top 7 gene set enrichment analysis (GSEA) of exclusive genes from CON, T3, and shared are depicted. Additional processes are shown in Supplementary file 5. (E) Sine curve was fitted for the selected clock genes. Gene expression of all groups was normalized by CON mesor. (F) For mesor, amplitude, and phase delta assessment, CircaCompare algorithm was used. The CON group was used as baseline. Additional genes (Per3, Rorc, Tef, Hif1a, and Nfil3) were used for these analyses. One-sample t-test against zero value was used and only phase was different from zero (mean 1.036, p<0.001). n = 4 samples per group and timepoint, except for the T3 group at Zeitgeber time (ZT) 22 (n = 3).