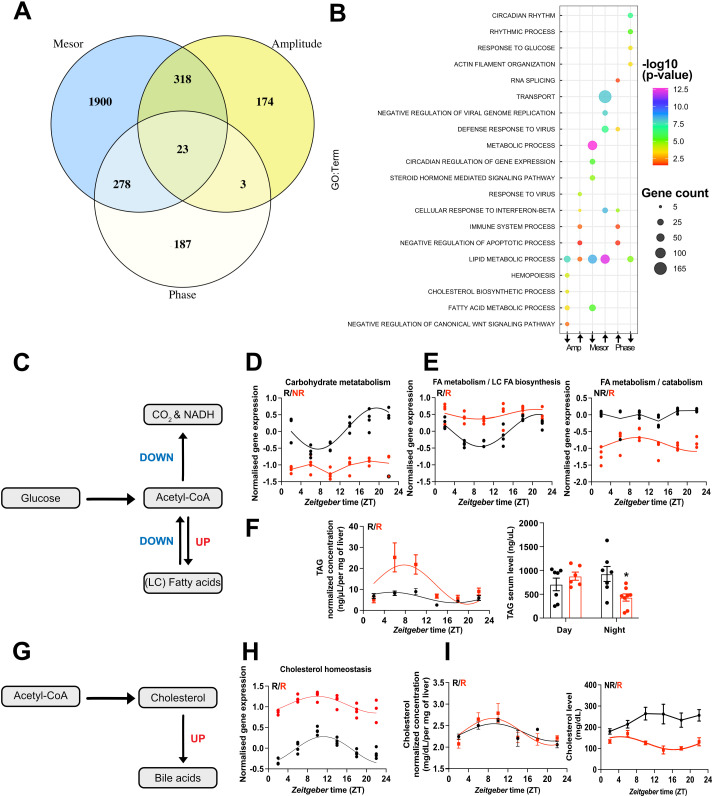
Figure 5. CircaCompare analyses of triiodothyronine (T3) (red) mice compared to control (CON) (black).
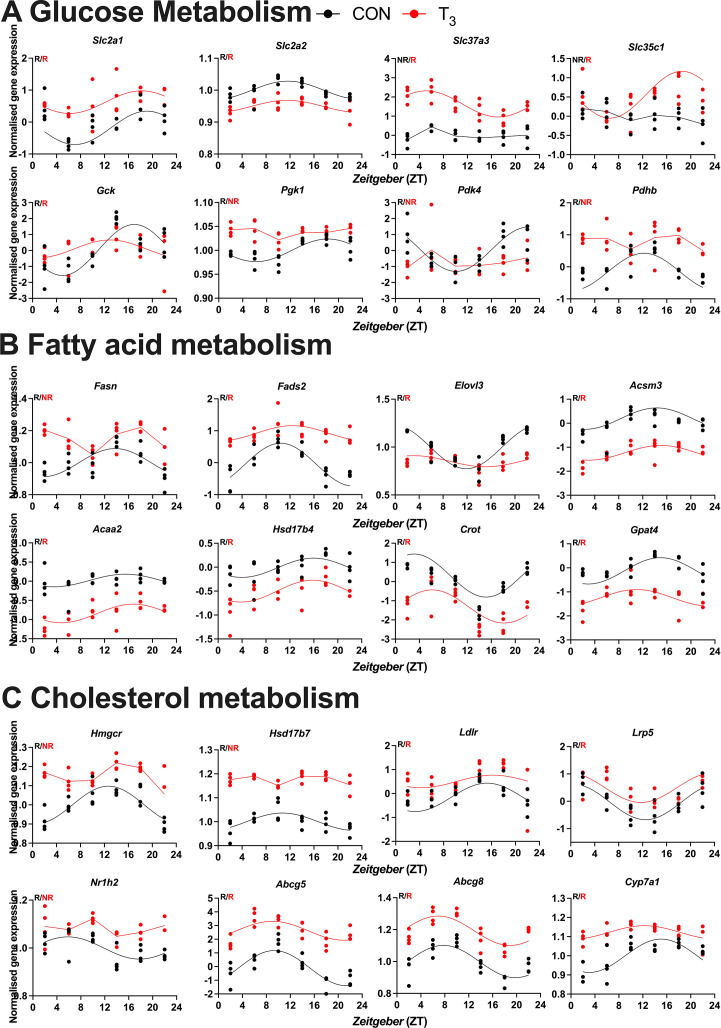
(A) Venn diagram demonstrates the number of probes that displayed differences in each rhythmic parameter (mesor, amplitude, and phase). (B) Top 5 enriched biological processes for each rhythmic parameter category. (C) Summary of the CircaCompare analyses regarding glucose and fatty acid (FA) metabolism. (D, E) Representation of glucose and FA metabolism biological processes obtained from transcriptome data. (F) Diurnal rhythm evaluation of liver triacylglyceride (TAG) and day (Zeitgeber time [ZT] 2–6) vs. night (ZT18–22) serum TAG levels comparisons. (G) Summary of the CircaCompare analyses regarding cholesterol metabolism. (H) Representation of cholesterol homeostasis obtained from transcriptome data. (I) Diurnal rhythm evaluation of liver and serum cholesterol. Gene expression from each biological process was averaged per ZT and plotted. The reader should refer to the text for detailed information regarding the changes found at the gene level of these processes. Sine curve was fitted for each rhythmic biological process. Individual gene expression pertaining to these processes is found in Figure 5—figure supplement 1. n = 4 samples per group and timepoint, except for the T3 group at ZT 22 (n = 3).