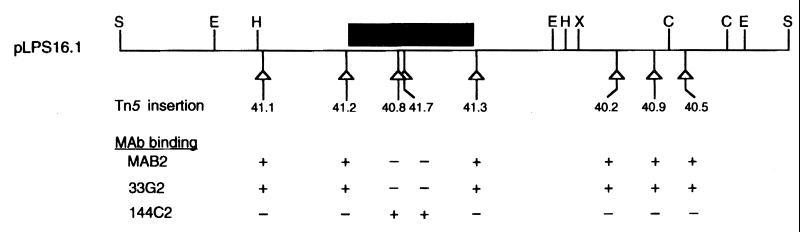
FIG. 3.
Localization of lag-1 by Tn5 mutagenesis. Tn5 insertions were introduced into pLPS16.1 and mapped according to the methods of de Bruijn and Lupinski (2). Each triangle represents the position of a Tn5 insertion in pLPS16.1. Plasmids containing each of the Tn5 insertions were electroporated into strain CS332, and transformants were tested for the ability to bind MAB2, 33G2, and 144C2. (−), insertions eliminating binding of MAB2 and 33G2; (+), insertions having no effect on the binding of MAB2 and 33G2. Wild-type LPS has the following MAb binding pattern: MAB2 (+), 33G2 (+), 144C2 (−). Restriction sites: C, ClaI; E, EcoRI; H, HindIII; S, SphI; X, XhoI. The black box defines the physical location of the lag-1 locus as determined by DNA sequencing experiments.