Figure 4.
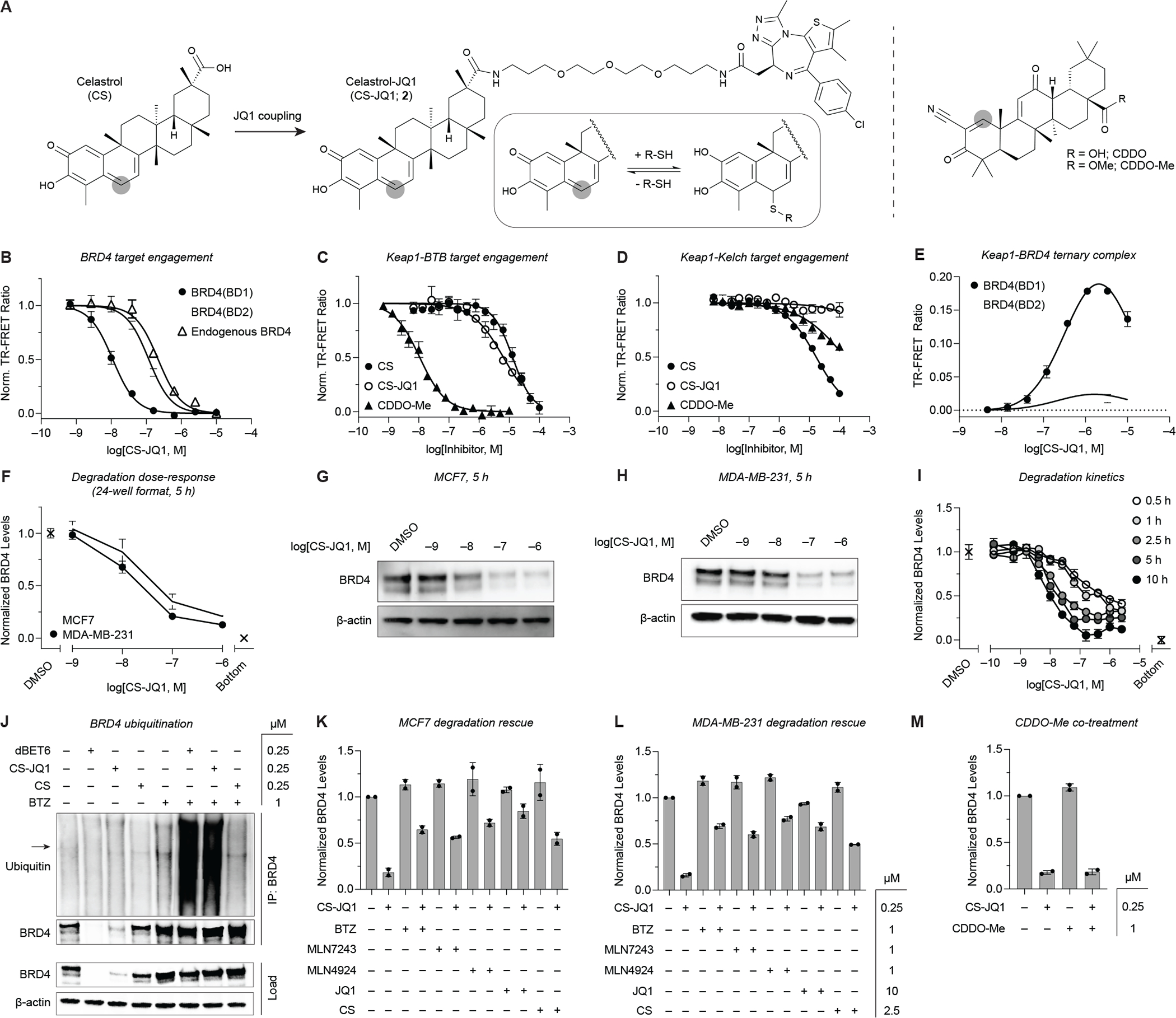
Celastrol is an E3 ubiquitin ligase recruiter suitable for PROTAC applications. (A) Chemical structures of CS, CS-JQ1 (2), CDDO, and CDDO-Me. Thiophilic sites are highlighted with grey circles. (B-D) Target binding of the various compounds for (B) recombinant BRD4(BD1), BRD4(BD2), and endogenous BRD4 in MCF7 cell extract (n = 2), (C) Keap1-BTB domain (n = 4), and (D) Keap1-Kelch domain (n = 2) (see STAR Methods). (E) CS-JQ1-induced ternary complex formation between full-length Keap1 and individual BRD4 bromodomains (n = 2). (F) TR-FRET quantification of BRD4 levels in MCF7 and MDA-MB-231 cells after treatment with a dose-titration of CS-JQ1 for 5 h in 24-well plates (n = 2). (G-H) Western blot analysis of the same samples used for TR-FRET quantification in (F). (I) Quantitative profiling of the time- and dose-dependent degradation of BRD4 in MDA-MB-231 cells by CS-JQ1 in 96-well plates (n = 2). (J) Ubiquitination of BRD4 induced by dBET6 and CS-JQ1 in MDA-MB-231 cells. Arrow in Ubiquitin blot represents BRD4 molecular weight (IP = immunoprecipitation). (K-L) Rescue of CS-JQ1-induced BRD4 degradation by BTZ, MLN7243, MLN4924, JQ1, and CS in (K) MCF7 or (L) MDA-MB-231 cells (n = 2). (M) Co-treatment of MDA-MB-231 cells with CS-JQ1 and CDDO-Me does not attenuate BRD4 degradation, indicating potential activity mediated through additional E3 ligase complexes other than Keap1/CRL3 (n = 2). Data in (B-E) are expressed as mean ± SD of n technical replicates and are representative of at least two independent experiments. Data in (F, I-M) are expressed as mean ± SD of 2 technical × 2 biological replicates. Unprocessed Western blot data is shown in Figure S4.
