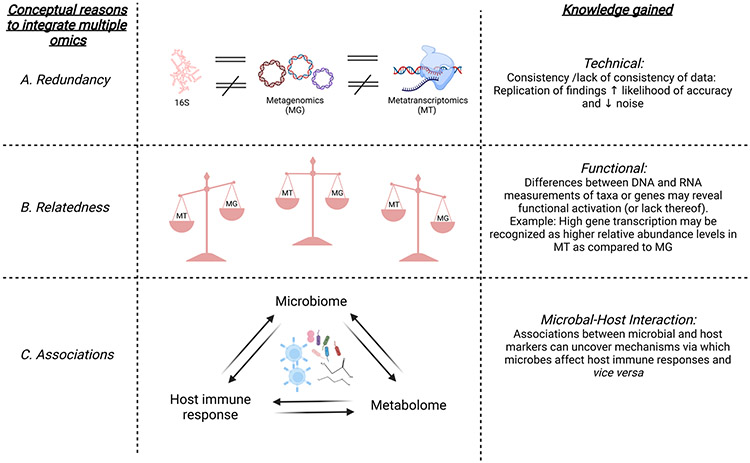
Figure 3: Inter-relatedness of various -omic approaches.
In this figure, we illustrate three different approaches to gain knowledge when combining multiple omic data: “redundancy”, “relatedness” and “associations”. A. Integration of omics helps to address “redundancy” by identifying consistency of data. In particular, replication of findings increases the likelihood of accuracy and decreases that of noise. B. The “relatedness” of the different omic methods has the potential to uncover differences related to function. As an example, high rates of gene transcription may be recognized as higher relative abundance in metatranscriptomics rather than in metagenomics for that specific gene, indicating possible functional activation. C. Finally, the integration of different omics can help discover the molecular mechanisms via which microbes affect host immune responses and vice versa.