Figure 1. In vivo CRISPR screen identifies negative transcriptional regulators of RGC survival and axon regeneration after ONC.
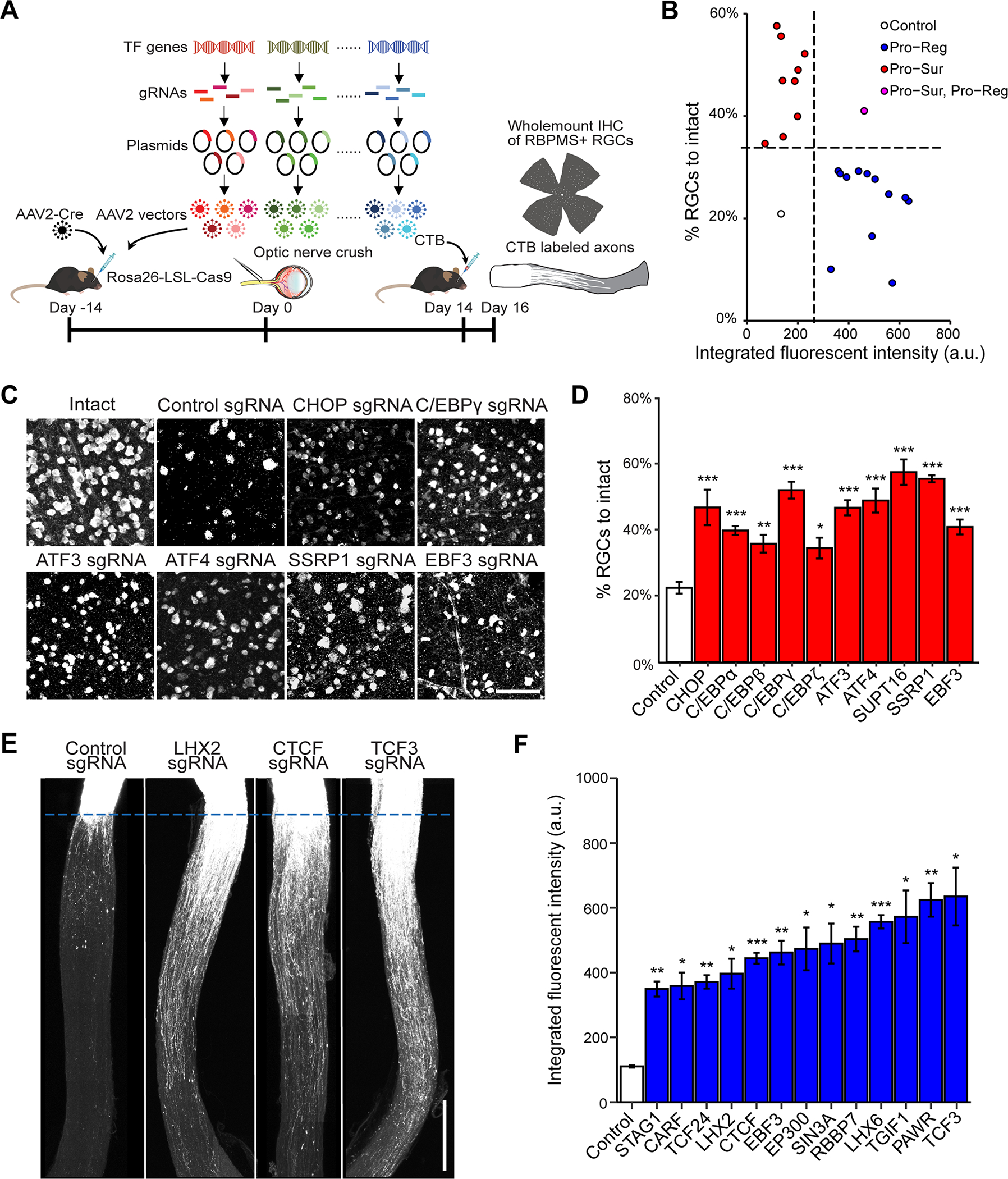
(A) Schematic illustration of the in vivo CRISPR screen in ONC model. AAV2 vectors encoding sgRNAs were injected intravitreally to Rosa26-LSL-Cas9 mice at 2 weeks before ONC. To ensure ablation efficiency, the mix of five different sgRNAs (from GeCKO mouse v2 CRISPR knockout library) were selected to target each gene candidate.
(B) The final lists of survival and regeneration hits were categorized into three groups: (1) deletion of the TF increasing axon regeneration without affecting RGC survival (Pro-Reg, 12 hits); (2) deletion of the TF increased RGC survival without affecting axon regeneration (Pro-Sur, 9 hits); (3) deletion of the TF promoted both RGC survival and axon regeneration (Pro-Sur, Pro-Reg, 1 hit). Each data point is the averaged results from RGC survival or axon regeneration analyses.
(C) Representative immunohistochemistry images of wholemount retinas showing improved survival after ONC injury by CRISPR ablation of individual TFs. Scale bar, 50 μm.
(D) Quantification of RGC survival with individual TF knockout. Data are shown as mean ± s.e.m. with n = 4–5 biological repeats. *p<0.05, **p<0.01, ***p<0.001, calculated by one-way ANOVA.
(E) Representative optic nerve images showing axon regeneration after ONC injury with individual TF knockout. Scale bar, 0.5 mm.
(F) Quantification of CTB labeled fluorescent intensity (from crush site) for all sgRNA hits that promote RGC axon regeneration. Data are shown as mean ± s.e.m. with n=5–6 biological repeats. *p<0.05, **p<0.01, ***p<0.001. Welch anova test, followed by Dunnett’s T3 adjustment.
