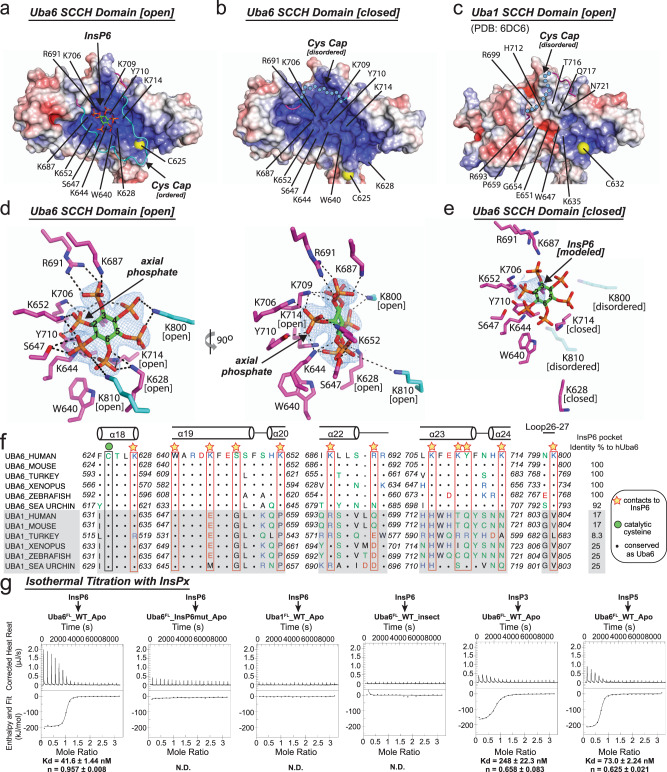
Fig. 3. InsP6 binds to a highly basic pocket unique to the SCCH domain of Uba6.
a–c Surface electrostatic representations of the SCCH domains of human Uba6 open (a) Uba6 closed (b) and Uba1 (c) structures. Uba6 residues involved in contacts to InsP6 in the Uba6 open structure and the corresponding residues from the Uba6 closed and Uba1 structures are labeled. The ordered cys cap in the Uba6 open structure is shown as cartoon and colored cyan and the disordered cys caps of Uba6 closed and Uba1 are shown as dashed cyan circles. d Composite omit density map (contoured at 1σ) for InsP6 in the Uba6 open structure is shown as blue mesh. InsP6 is shown as sticks with carbons (green), oxygens (red), phosphates (orange). InsP6-interacting residues are shown as sticks and hydrogen bonds are shown as dashed lines. e Composite omit density map for InsP6 binding pocket in the Uba6 closed structure is shown as in d. InsP6 has been modeled based on the Uba6 open structure. f Structure-based sequence alignment of InsP6 binding region in Uba6 and Uba1 SCCH domain. Secondary structure for Uba6 is shown above. Residues important for InsP6 binding are highlighted by yellow stars. g Isothermal titration calorimetry data for interactions between the indicated Uba6 and Uba1 variants with the indicated inositol phosphate. Experiments were performed in triplicate and upper panels show raw power data and lower panels show fits of the data to standard binding equations using NanoAnalyze software (TA instruments). Throughout the figure, “Apo” labels refer to E. coli-derived material.