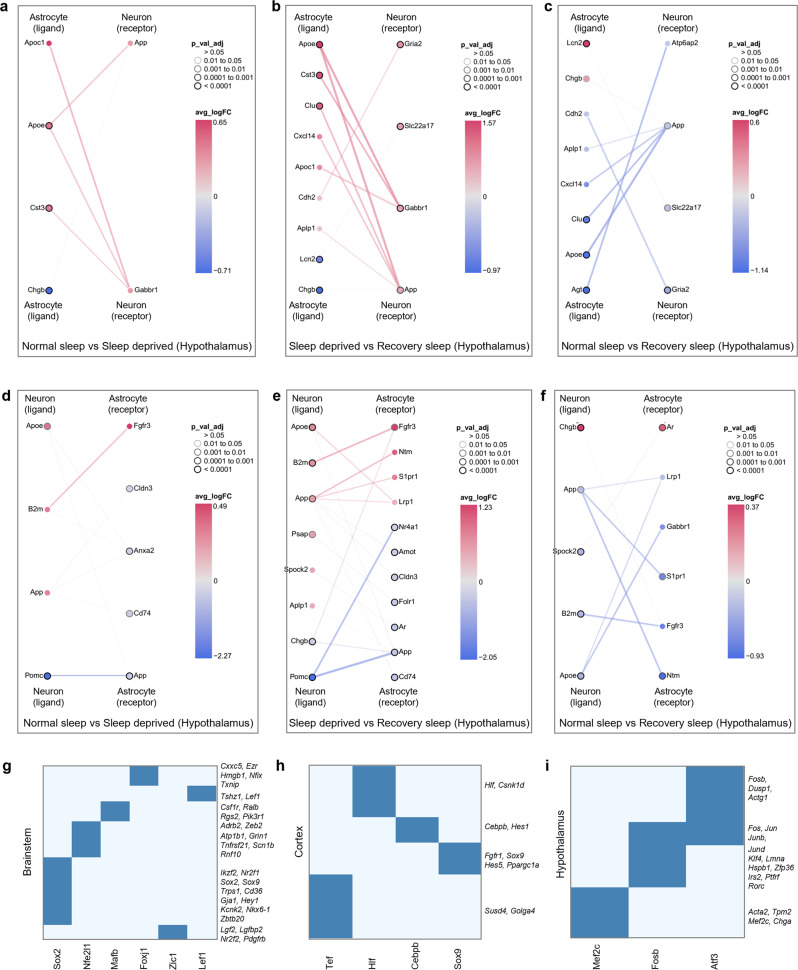
Fig. 7. Sleep need modulates cell-cell communication and gene regulatory networks.
a–f Interactions between differentially expressed ligands of astrocytes and receptors of neurons for (a) NS versus SD, (b) SD versus RS, and (c) NS versus RS comparisons, and ligands of neurons and receptors of astrocytes for (d) NS versus SD, (e) SD versus RS, and (f) NS versus RS comparisons in the hypothalamus. Nodes represent ligands or receptors in astrocytes and neurons, respectively. Node outline thickness indicates the level of significance (Benjamini-Hochberg adjusted P-value). Colour of nodes and edges represents the magnitude of alteration in expression (log fold-change, logFC) (see Methods). g–i Uniquely expressed transcription factors (rows) in sleep-deprived cells and their co-expressed gene sets (columns) in (g) brainstem, (h) cortex, and (i) hypothalamus. For specific GO terms of the co-expressed genes, see Supplementary Table 5. NS normal sleep, SD sleep deprived, RS recovery sleep.