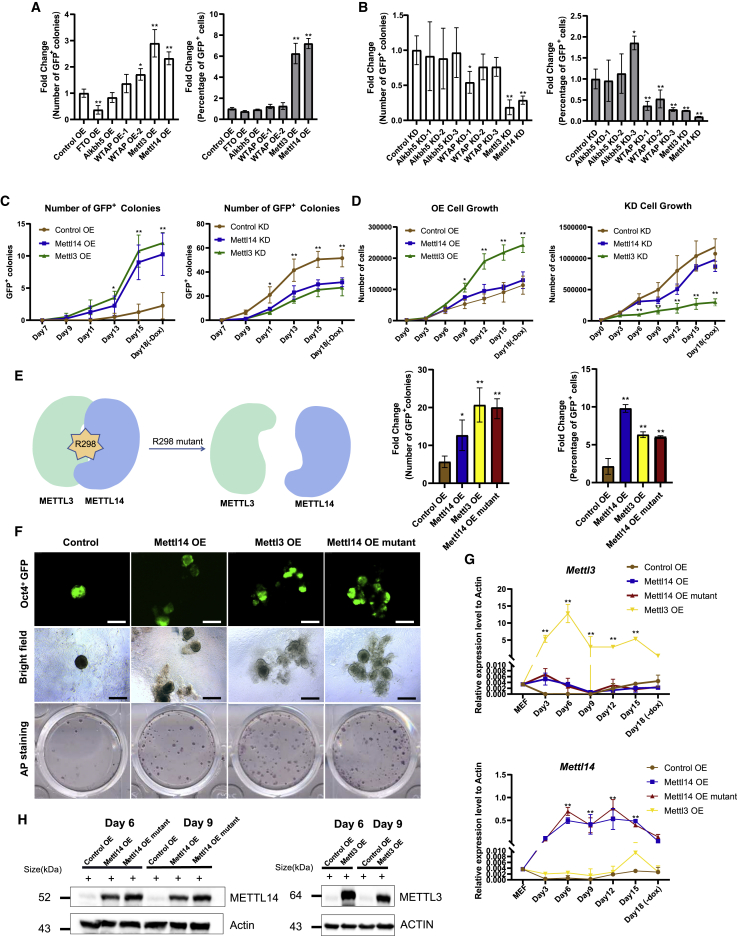
Figure 1.
Mettl14 can facilitate reprogramming in an m6A-independent manner
(A) The number of Oct4-GFP+ colonies was counted, and the percentage of Oct4-GFP+ cells in the overexpression (OE) group was analyzed by FACS 18 days after induction (starting MEF density was 8,000 cells/well in a 12-well plate). The data are presented as average fold change of Oct4-GFP+ colonies (left panel) or percentage of Oct4-GFP+ cells (right panel) ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student’s t test performed for comparison (control OE, empty vector control).
(B) The number of Oct4-GFP+ colonies was counted, and the percentage of Oct4-GFP+ cells in the knockdown (KD) group was analyzed by FACS 18 days after induction (the MEF starting density was 12,000 cells/well of a 12-well plate). The data are presented as average fold change of Oct4-GFP+ colonies (left panel) or percentage of Oct4-GFP+ cells (right panel) ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student’s t test performed for comparison (control KD, scramble short hairpin RNA [shRNA] control).
(C) The number of Oct4-GFP+ colonies formed was facilitated by Mettl14 or Metttl3 OE. The opposite effect was observed after the expression of each was knocked down. The MEF starting density was 8,000 cells/well for OE and 12,000 cells/well for KD in a 12-well plate. The data are presented as the means ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student’s t test performed for comparison.
(D) Cells were counted at different time points during reprogramming, and growth curves were plotted. The data are presented as the means ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student’s t test performed for comparison.
(E) Schematic representation of the mutation at the Mettl14 R298E locus (left panel). Estimated reprogramming efficiency of R298E mutant-expression cells as determined by the number of Oct4-GFP+ colonies formed and the percentage of Oct4-GFP+ cells (middle and right panels) (the MEF starting density was 6,000 cells/well in a 12-well plate). The data are presented as the means ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student’s t test performed for comparison.
(F) Morphology of the Oct4-GFP+ primary colonies (top and middle panels). Representative image of AP-stained plates captured 18 days after induction (bottom panel). Scale bars, 400 μm.
(G) qRT-PCR analysis showing the expression level of Mettl3 and Mettl14 in the iPSCs at RNA levels. The data are presented as the means ± SEM (n = 3); ∗p < 0.05, ∗∗p < 0.01 by Student’s t test performed for comparison.
(H) Western blot showing the expression level of Mettl3 and Mettl14 in the iPSCs at protein levels. ACTIN is used as loading control.