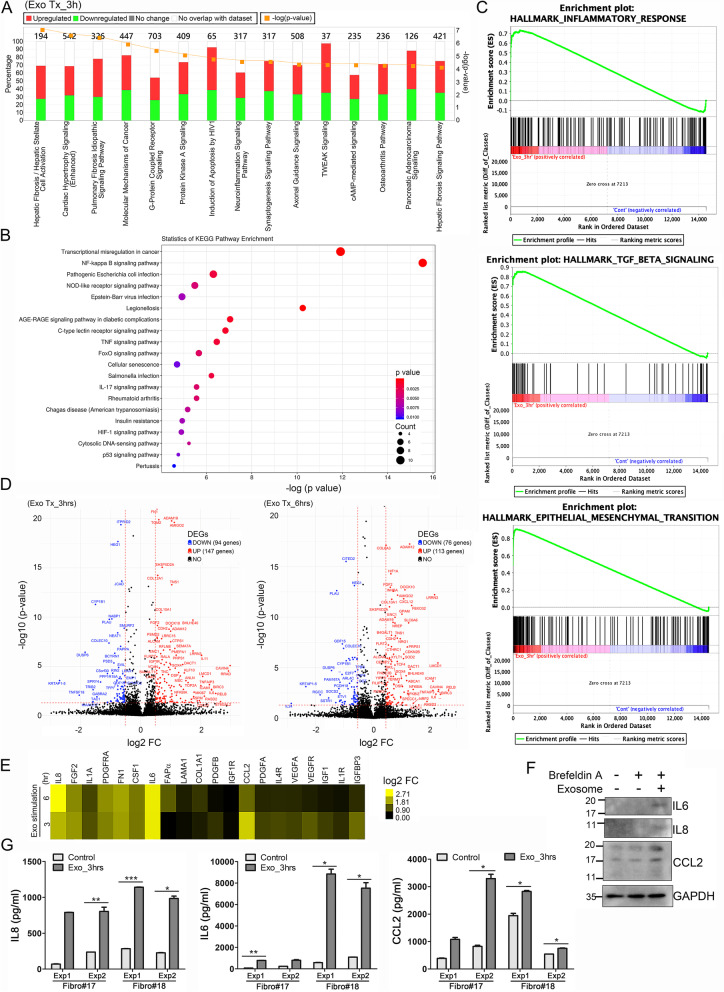
Fig. 2.
EBV product-containing exosomes enhance fibrotic and inflammatory signalings in fibroblasts. A Analysis of signaling pathways in fibroblasts stimulated with HK1EBV cell-derived exosomes compared with those exposed to medium. The top-scoring signaling pathways were identified using Ingenuity Pathway Analysis (IPA) software (QIAGEN). The identified pathways were ranked by significance [−log (p-value)]. The number of genes that met cutoff criteria [|log2(FC)| > 0.5 and p value < 0.05] in a given signaling pathway was shown. Total RNAs were harvested 3 hours post HK1EBV cell-derived exosome (10 μg/ml) stimulation. B Analysis of KEGG pathways in fibroblasts stimulated with HK1EBV cell-derived exosomes compared with those exposed to medium. C Gene set enrichment analysis (GSEA) in fibroblasts treated with HK1EBV cell-derived exosomes. GSEA plots for gene sets involved in inflammatory response, TGFβ signaling, and epithelial-mesenchymal transition were identified. Expression values were normalized to that of control fibroblasts. Significance threshold set at FDR < 0.25. D Volcano plots of transcriptomic analyses revealed differentially expressed genes (DEGs). Cutoff values were defined as |log2(FC)| > 0.5 and a P-value < 0.05. Total RNA was harvested 3 hours and 6 hours post exosome stimulation (10 μg/ml). E Expression heatmap of select genes upregulated by EBV product-containing exosomes. Expression levels were measured by qRT-PCR and normalized with respect to peptidylprolyl isomerase A (PPIA), used as an internal control. Values are expressed as log2 (fold change). F Western blot analysis of IL8, IL6, and CCL2 in fibroblasts. Protein lysates were harvested after 2-hour exosomal stimulation plus 4-hr treatment with Brefeldin A. GAPDH was used as a loading control. G Quantification of secreted IL8, IL6, and CCL2 in cell-free culture supernatants collected 3 hours post exosomal stimulation of fibroblasts. Data are presented as means ± SD of triplicate experiments (*p < 0.05**, p < 0.01, ***p < 0.001; paired t test)