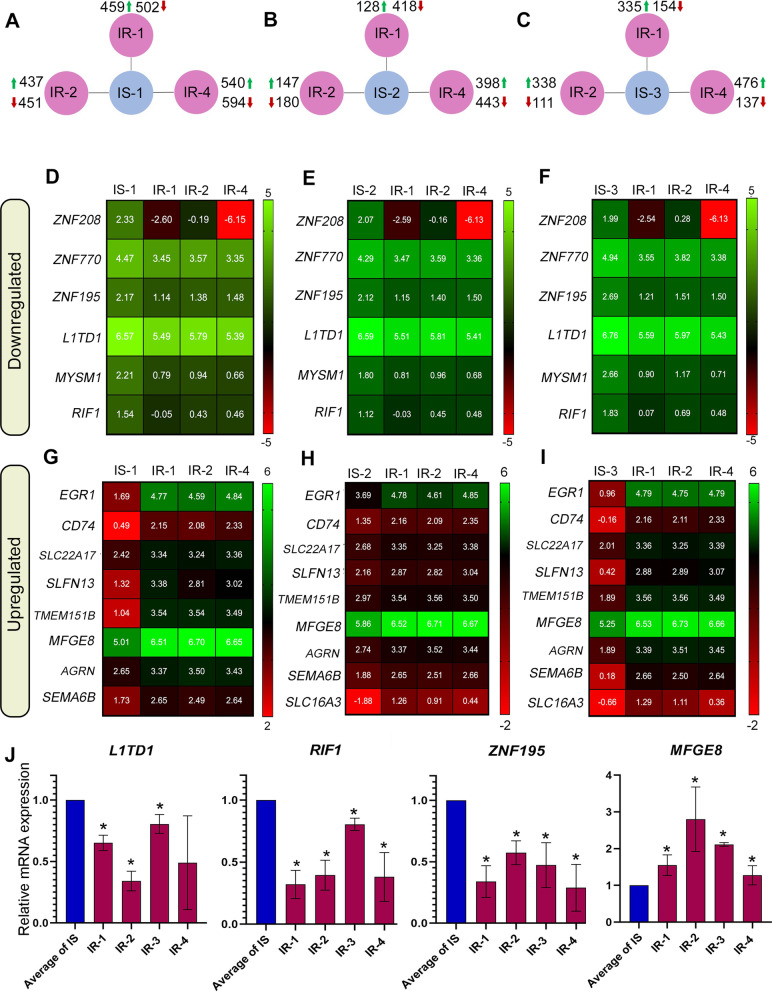
Fig. 3.
Transcriptomics of IS- and IR-iPSCs reveal differentially expressed genes (DEGs) in IR-iPSCs. A–C Summary of upregulated and downregulated DEGs for each comparison. Heat maps for D–F downregulated genes and G–I upregulated in all IR-iPSCs when compared individually to each of the IS-iPSCs based on RNA-Sequencing. Log2 of FPKM values of each IR-iPSC in comparison to IS-iPSC was plotted as heatmap. J Quantitative PCR results for select candidate genes identified from DEG analysis such as L1TD1, RIF1, ZNF195 and MFGE8 where delta CT for each IR-iPSC was normalized to the average delta CT for all IS-iPSC cell lines. (n = 2 clones), * P value < 0.05