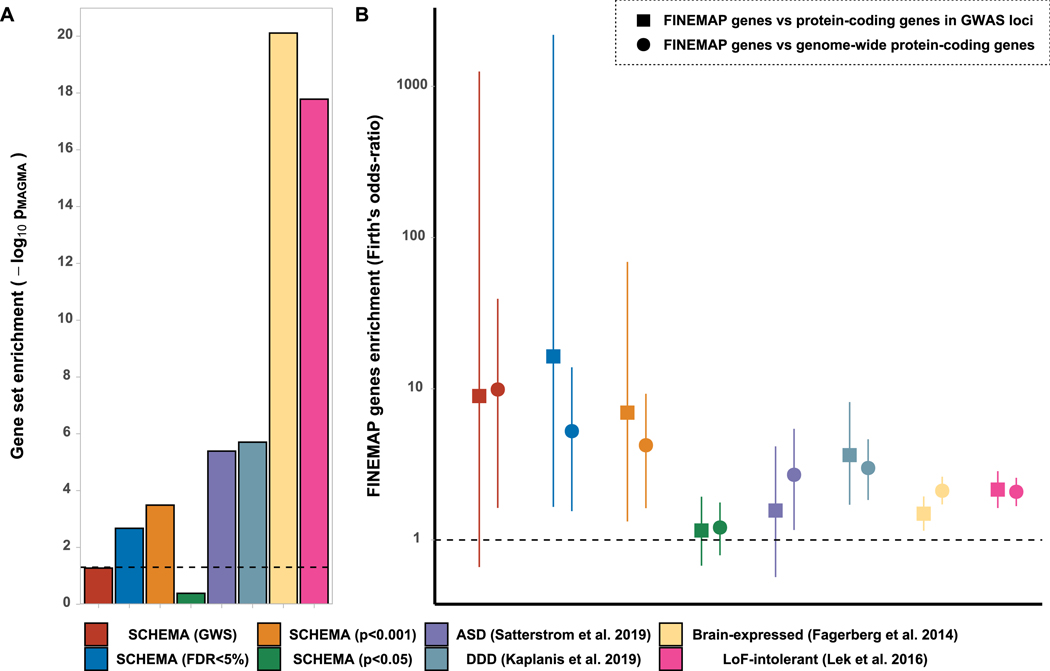
Figure 2: Gene set enrichment tests at genome-wide level and for protein coding genes containing FINEMAP credible SNPs.
Gene sets tested were retrieved from sequencing studies of schizophrenia (SCHEMA; companion paper), autism-spectrum disorder33 and developmental disorders32. Sets representing genes that are intolerant to loss-of function mutations40 (LoF-intolerant) and brain-expressed genes41are also shown. A) MAGMA gene set enrichment analysis, dotted line indicates nominal significance (p=0.05). B) Logistic regression (with Firth’s bias reduction method) showing the odds-ratio (and 95% CI) for association between protein-coding genes containing at least 1 credible FINEMAP SNP (N=418 after excluding genes with no LoF-intolerance data) and genes from the sets indicated. Odds-ratios are relative to protein-coding genes within GWAS K≤3.5 loci (1,283 genes, squares) or across the genome excluding the xMHC (19,547 genes; circles). Dotted line indicates no enrichment.