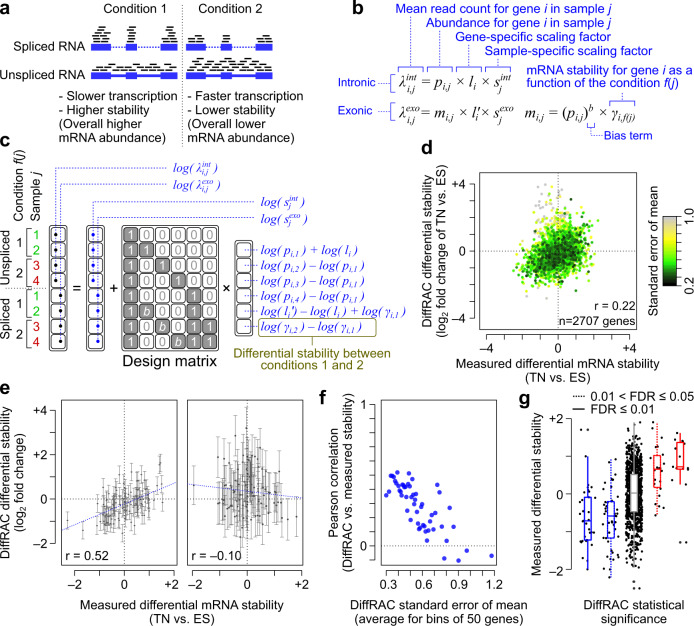
Fig. 1. Inference of differential mRNA stability using DiffRAC.
a Schematic representation of the effect of transcription and stability on the abundances of unspliced and spliced RNA. b DiffRAC models the mean (λ) of intronic (int) and exonic (exo) read distribution as a function of pre-mature (p) and mature (m) transcript abundances, in addition to gene-specific (l) and library-specific (s) scaling factors. Mature mRNA abundance is modeled as a function of the pre-mature RNA abundance and mRNA stability (γ), which is in turn a function (f) of the experimental variables. Also see Supplementary Fig. 1. c An example case with four samples and two experimental conditions, showing how DiffRAC’s model can be implemented in a regression with a log-link function, along with the interpretation of regression coefficients (also see Methods). d Comparison of DiffRAC stability estimates against experimental mRNA half-life (stability) measurements in mouse ES cells differentiated to terminal neurons (TN)15,18,19. Each data point stands for one gene, with the points coloured according the standard error of the mean (SEM) for DiffRAC estimates. e Comparison of DiffRAC estimates vs. measured mRNA stability for the 100 genes with the smallest (left) and largest (right) DiffRAC SEMs. Error bars represent the standard error of the mean (SEM). f The Pearson correlation between DiffRAC estimates and measured mRNA stability for bins of 50 genes sorted by their SEM. g Distribution of experimental mRNA half-life measurements for genes that DiffRAC has identified as significantly destabilized (blue boxplots) or stabilized (red boxplots) in TN vs. ES cells, at FDR cutoffs of 0.05 (dashed line) or 0.01 (solid line). Genes that are not called as significant by DiffRAC are represented with the grey boxplot.