Figure 4. Fur-regulated processes control the conditional essentiality of lipid A.
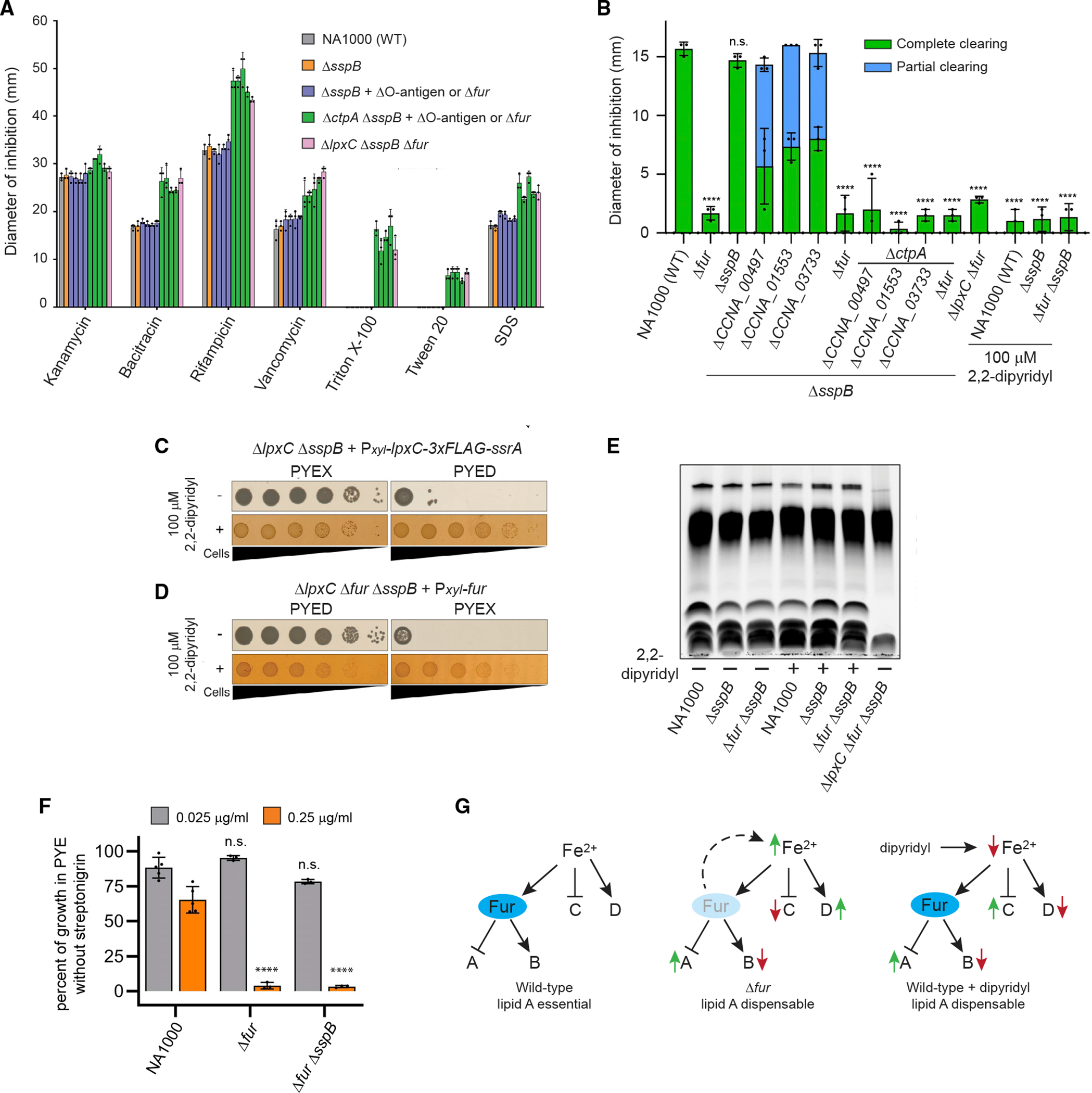
(A) Chemical sensitivity was measured by disc diffusion assay (mean ± SD). Dots indicate individual data points. Suppressor mutations present in strains represented by blue or green bars are, from left to right, ΔCCNA_00497, ΔCCNA_01553, ΔCCNA_03733, and Δfur.
(B) CHIR-090 sensitivity measured by disc diffusion assay (mean ± SD). Partial clearing indicates the diameter of a ring of intermediate growth. Dots indicate individual measurements, and significance was tested by one-way ANOVA followed by Dunnett’s post-test comparison with NA1000. ****p < 0.0001; n.s., not significant. Strains exhibiting rings of partial growth were excluded from the analysis.
(C) Viability of the LpxC depletion strain under inducing (PYEX) or depleting (PYED) conditions in the presence or absence of 100 μM 2,2′-dipyridyl.
(D) Viability of ΔlpxC Δfur ΔsspB cells harboring a Pxyl-fur plasmid, grown in noninducing (PYED) or inducing (PYEX) conditions in the presence or absence of 100 μM 2,2′-dipyridyl. Plates included kanamycin to retain the expression vector. Brightness was reduced and contrast increased to improve the clarity of colonies grown on 2,2′-dipyridyl.
(E) Proteinase K-treated lysates of the indicated strains grown overnight in the presence or absence of 100 μM 2,2′-dipyridyl. Samples were normalized by OD660.
(F) Growth inhibition by SNG in liquid PYE cultures of the indicated strains (mean ± SD). Dots represent individual OD660 ratios, and significance was tested by one-way ANOVA followed by Šídák’s multiple comparisons test, where each strain was compared with NA1000 grown under the same condition. ****p < 0.0001; n.s., not significant.
(G) Changes in Fur-regulated gene expression correlate with the ability to survive in the absence of lipid A. Genes regulated by Fur in concert with iron (sets A and B) are modulated similarly by deletion of fur or by iron limitation, whereas genes regulated by iron alone (sets C and D) are modulated in opposite directions in these two conditions.
