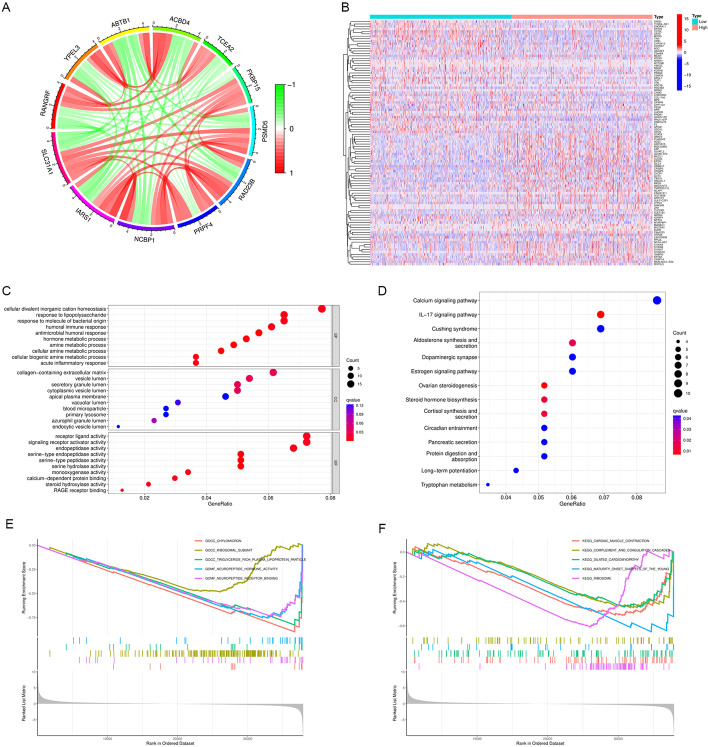
Fig. 6.
Identification of related genes, pathways and cellular functions of SLC31A1. A Circos graph displaying the co-expression networks of SLC31A1 with 11 genes in breast cancer samples. Each sector of the circle represents one gene, and its width indicates the total amount of co-occurrence that connects one certain gene to the other. The width of each link represents the total co-expression times of the linked genes. B Heatmap showing 186 down-regulated genes (blue) and 164 up-regulated genes (red) identified in the high expression group. The colored matrix shows hierarchical clustering of expression microarrays, with each column representing the log2 (TPM) values of expression levels of 350 genes in microarray hybridization of one sample, and each row representing the expression of a particular gene described to the right. C Results from Gene Ontology (GO) analysis. Genes of the immune response and metabolic process were mostly enriched. D Results from Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Genes of the calcium signaling pathway and IL-17 signaling pathway were mostly enriched. E GSEA analysis revealed down-regulated pathways associated with SLC31A1 expression. The GSEA analysis of GO between the high SLC31A1 expression samples and low SLC31A1 expression samples revealed significant difference in the enrichment of chylomicron and triglyceride pathways. F The GSEA analysis of KEGG pathway enrichment between the high SLC31A1 expression samples and low SLC31A1 expression samples revealed significant difference in the enrichment of dilated cardiomyopathy, cardiac muscle contraction and ribosome pathways