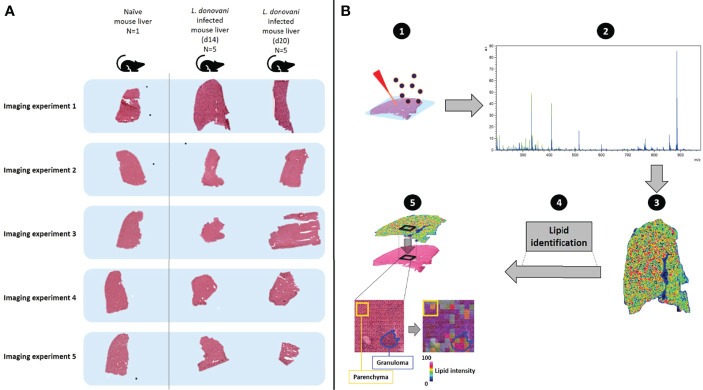
Figure 1.
Schematic describing workflow for monitoring spatially-resolved lipids during hepatic Leishmania donovani infection. (A) Livers were obtained from a naïve C57BL/6 mouse and from L. donovani-infected C57BL/6 mice at d14 and d20 p.i. (n=5 mice per time point). Samples were sequentially measured, using the naïve tissue as a baseline and comparator (n=1). In addition, consistency of naïve mouse livers (n=5) is shown in Figure S1. (B) The spatial distribution of lipids and their respective structural identification was assessed by MALDI-MSI (1): a chemical matrix was applied to the tissue surface – this absorbs lipids from the tissue while retaining spatial localization. Subsequently, lipids were ionized by MALDI, which generates charged ions in the gas-phase (2). A mass analyzer is used to determine the mass-to-charge (m/z) ratio of these ions from which (3) the ions can be visualized to reveal their spatial localization and relative abundance across the analyzed tissue (4). Structural identification of spatially- resolved lipids was performed by tandem mass spectrometry (5). H&E-stained tissues were co-registered with mass spectrometry images, to allow integration of molecular lipid information from annotated regions of interest (i.e. granulomas and surrounding parenchyma) with histopathology.