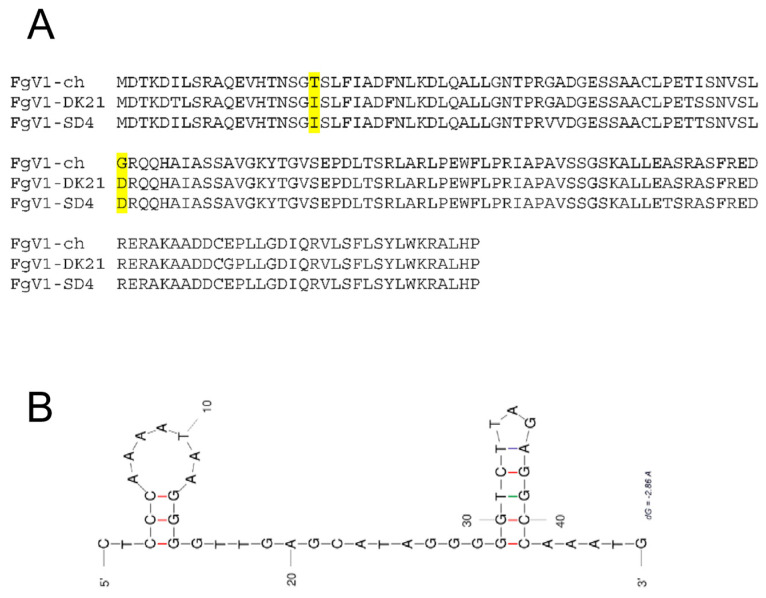
Figure 2.
Amino acid changes in FgV1 ORF2-encoded protein (pORF2) alignment and predicted 3′-UTR secondary structure of FgV1-SD4 were speculated to explain the hypovirulence effects. (A) Amino acid alignment of pORF2 among FgV-1-ch, FgV1-DK21, and FgV1-SD4. (B) The predicted 3′-UTR (CTCCCAAAATAAGGGGTTGAGCATAGGGGGTCTTAGAGGCCAAATG) secondary structure of FgV1-SD4 with hairpin loops (UNAfold, DINAMelt quickfold, and mfold); initial ΔG = −9.00. The highlighted amino acid residues indicate the two residues different among the three reported viral variants which could be associated with different levels of hypovirulence observed.