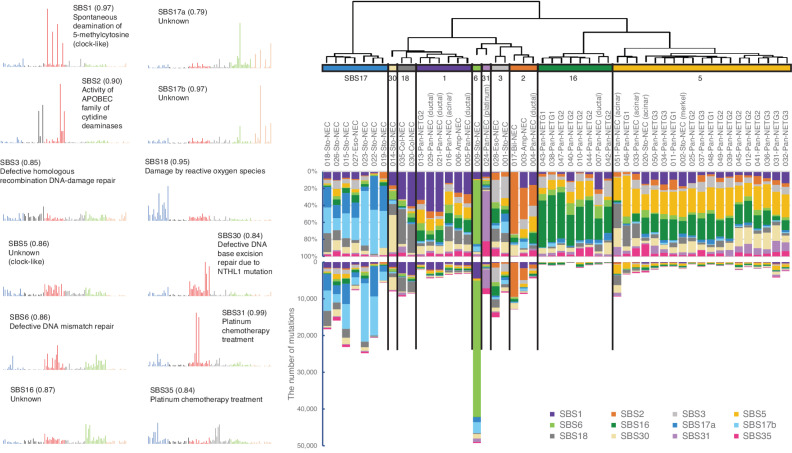
Figure 4.
Mutational signature analysis of GIS-NENs. Hierarchical clustering by de novo extraction of mutational signatures with nonnegative matrix factorization of available WGS data. Stability plotting indicated 12 mutational signatures (>0.85; Supplementary Fig. S18). Their profiles and functions were assigned based on COSMIC SBS Signatures (v3.1). Sto, stomach; Eso, esophagus, Col, colorectum; Pan, pancreas; Amp, ampullary; Bil, bile duct; Platinum, platinum chemotherapy treatment; Merkel, gastric NEC with Merkel cell polyomavirus. The parentheses in the left figure indicate cosine similarity to COSMIC signatures.