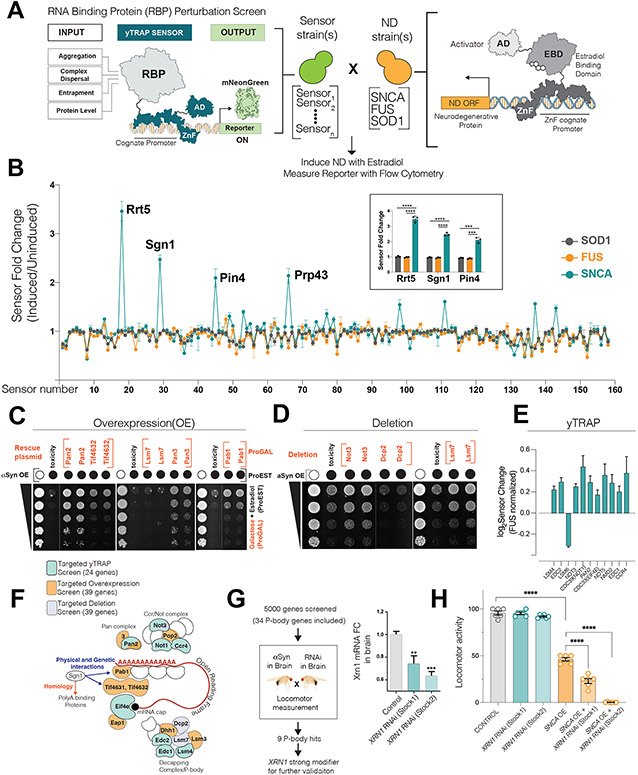
Figure 1. Processing body genes are modifiers of αS mediated toxicity in yeast and Drosophila.
(A) Logic of RBP perturbation screen. The RBP-yTRAP platform (left) is crossed to ND protein expressing yeast (right). Upon induction of ND protein, the mNeonGreen output signal was quantitated by flow cytometry. ZnFs for ZEM module and yTRAP module are different. (AD: Activator domain, ZnF: ZincFinger, RBP: RNA-binding protein, ND: neurodegeneration)
(B) Perturbation screen results. Y axis: Fluorescence linear fold change (induction of ND protein versus no induction), X-axis: RBP sensors (3 biological replicate screens; mean, sd). Four of the most perturbed proteins upon αS induction are marked and the inlet shows the graph for three of these proteins (mean, sd, t-test, ***p<0.001, ****p<0.0001).
(C, D, E) αS toxicity modifier mini-screens guided by Sgn1 yTRAP signal. 39 genes for overexpression/deletion and 24 genes for yTRAP screen. (C, D) Representative plate growth assays of αS toxicity modifiers. (Dotted line indicate a cut within the same plate). (E) yTRAP assay showing αS specific significant perturbation signals (in comparison to Fus) for 11 genes. (n=4 biological replicates, mean, sd, p<0.0005 for all 11 genes, multiple comparison corrected among 24 tested genes). For all the 24 genes tested, see Supp.Fig1H.
(F) Sgn1 guided mini-screens for αS toxicity modifiers converge on complexes within P-body granules. Miniscreens are color-coded.
(G-H) Locomotor based forward genetic screen in flies.(G) RT-qPCR validation of XRN1 knockdown in brains of two RNAi stocks (n=6, mean± sem, two-tailed t-test). (H) Locomotor activity measurements (mean± sd) of flies. Data were analyzed using one-way ANOVA with Tukey post-hoc analysis (n=6).