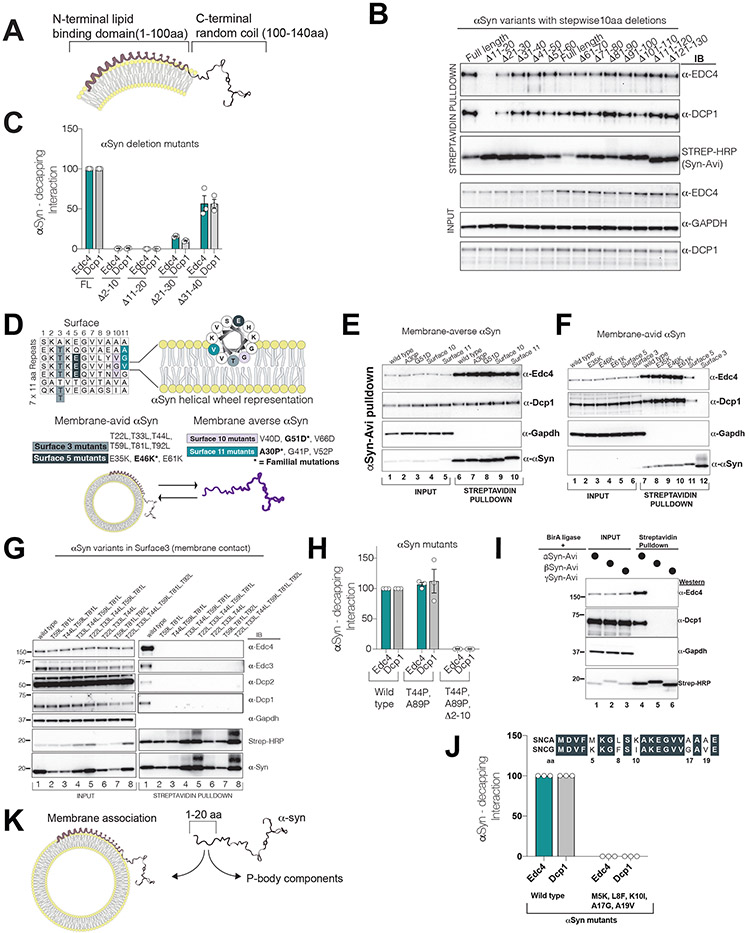
Figure 3. The N-terminus of αS dichotomously interacts either with decapping-module proteins or with membranes.
All experiments were performed by transient transfection of a single plasmid containing αS-Avi mutant co-expressing BirA ligase in HEK293 cells. αS pulldown was performed by Streptavidin beads and probed for P-body proteins.
(A) A schematic of αS on a bilayer membrane with its acidic C-terminus projecting away from the membrane.
(B) 10 aa deletion-scan of αS-Avi in HEK293 cells in lipid binding domain(1-100 aa). Note the absence of P-body interactions in Δ11-20 aa mutant.
(C) The first 20 aa residues of αS are essential for decapping module interactions (quantified interactions, n=3, mean,sem).
(D) Logic of membrane-averse or membrane-avid stratification of αS surface mutants. Up: αS 11 aa imperfect repeats and the helical view of αS on the right. Below: Familial point mutations in different surfaces are bolded. Combined surface mutants are rationally designed to amplify the effect of the single familial mutation.
(E) Membrane-averse αS G510D and A30P familial mutations (and their extended surface mutants 10 and 11, respectively) do not affect P-body interactions.
(F) Membrane avid αS mutants abrogate P-body interactions (see surface3 and 5).
(G) Increasing membrane avidity of αS by mutating Surface3 threonines to leucine completely diminish decapping module binding.
(H) Non-membrane-bound αS mutant (T44P and A89P) interacts with decapping module interactions through its first 10 aa.
(I) Among synuclein family members, only αS interacts with P-body components.
(J) Replacement of gamma synuclein residues in the first 20aa to alpha synuclein impairs decapping binding.
(K) Proposed model for dichotomous interaction of αS with membranes or P-body components.