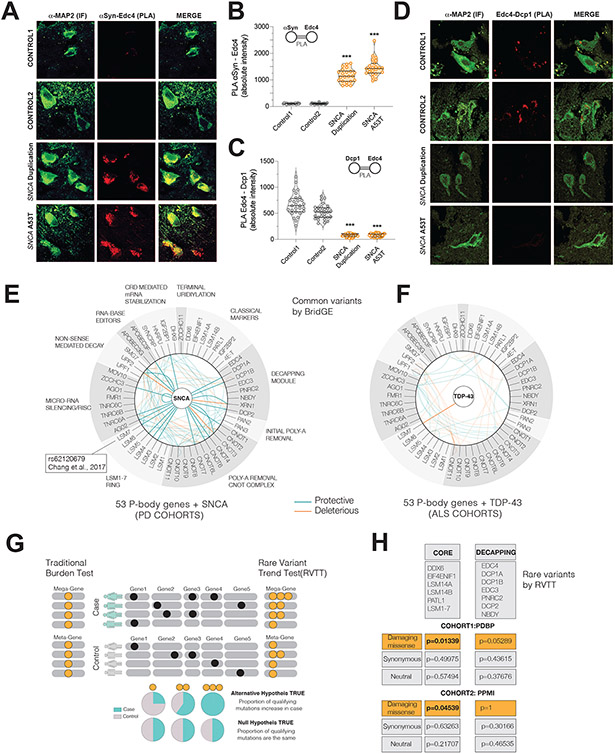
Figure 7. Postmortem brain and human-genetic analyses further connect P-body perturbations to PD.
(A,B) PLA between Edc4-αSyn in PD patient and control brains and its quantification(D) (*** p<0.001, one way ANOVA with Bonferroni correction, Map2 neuronal marker)
(C,D) PLA between Edc4-Dcp1 in PD patient and control brains and its quantification(E) (*** p<0.001, one way ANOVA with Bonferroni correction)
(E-F) Common variant BridGE analysis in PD and ALS cohorts to infer putative gene-gene interactions among P-body genes and SNCA. 53 P-body genes are grouped according to their functions. At the center, the causal genes are shown, SNCA for PD and TDP-43 for ALS. The map of significant SNP interactions (summarized at the gene level) with P-body genes and TDP-43 (TARDBP) (G) and SNCA (H) are shown. Protective and deleterious interactions are color coded. In (H), the LSM7 SNP GWAS genome-wide hit is indicated. Reduced opacity: The intra P-body interactions, High opacity: SNCA and P-body gene interactions.
(G) Logic of RVTT analysis. Left: Traditional Burden test collapse all mutations irrespective of their frequency. Right: RVTT accounts for trends in qualifying-mutation proportions in case versus controls. Mega-gene represents all genes in the pathway. Orange dots are collapsed qualifying mutations.
(H) Rare Variant Trend test (RVTT) analysis of P-body pathway genes In two different PD cohorts, PPMI and PDBP, RVTT was applied to P-body genes. Two “bags” of P-body genes were tested: CORE genes and DECAPPING genes, which are indicated in the lists. Three separate mutation-types (missense damaging, missense neutral and synonymous) were interrogated in both cohorts.