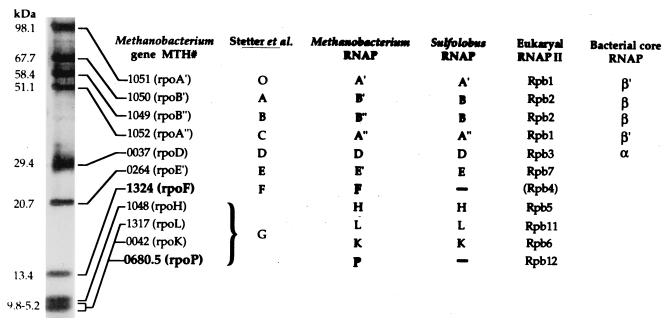
FIG. 1.
Identities of M. thermoautotrophicum ΔH RNAP subunits silver stained after separation by tricine-SDS-PAGE. Stetter et al. (24) identified eight subunits (designated by the letters O and A through G) in RNAP preparations from M. thermoautotrophicum Winter. Based on estimated sizes and very similar patterns of SDS-PAGE resolution, O and A through F appear to have been homologs of the M. thermoautotrophicum ΔH subunits here designated A′, B′, B", A", D, E′, and F, and subunit G was a mixture that contained the four polypeptides designated subunits H, L, K, and P. With the exception of subunits F and P, the M. thermoautotrophicum ΔH subunits are homologs of subunits identified previously for Sulfolobus acidocaldarius RNAP (14, 15). M. thermoautotrophicum ΔH RNAP preparations did not contain a homolog of the S. acidocaldarius subunit G, and the MTH0265 and MTH0040 gene products, predicted to be RNAP subunits E" (6.7 kDa) and N (6.4 kDa) (23), were not detected, although their presence may have been masked within the cluster of similarly sized, small subunits. Eucaryal and bacterial homologs of the M. thermoautotrophicum ΔH subunits are listed based on motif conservation and sequence alignments (5, 16, 21, 22). Members of the MTH1324 family (subunit F) have limited similarity to eucaryal Rpb4, a nonessential subunit of RNAPII that in yeast participates in transcription under stress conditions and at temperature extremes (4, 20). All members of the MTH0680.5 family (subunit P) contain four cysteinyl residues arranged in a manner consistent with a C-4 zinc finger motif and homology to Rpb12 (see Fig. 2C). .