Figure 5. MAPK inhibitor-induced changes in cJUN and p-cJUN levels correlate with drug-induced changes in differentiation states.
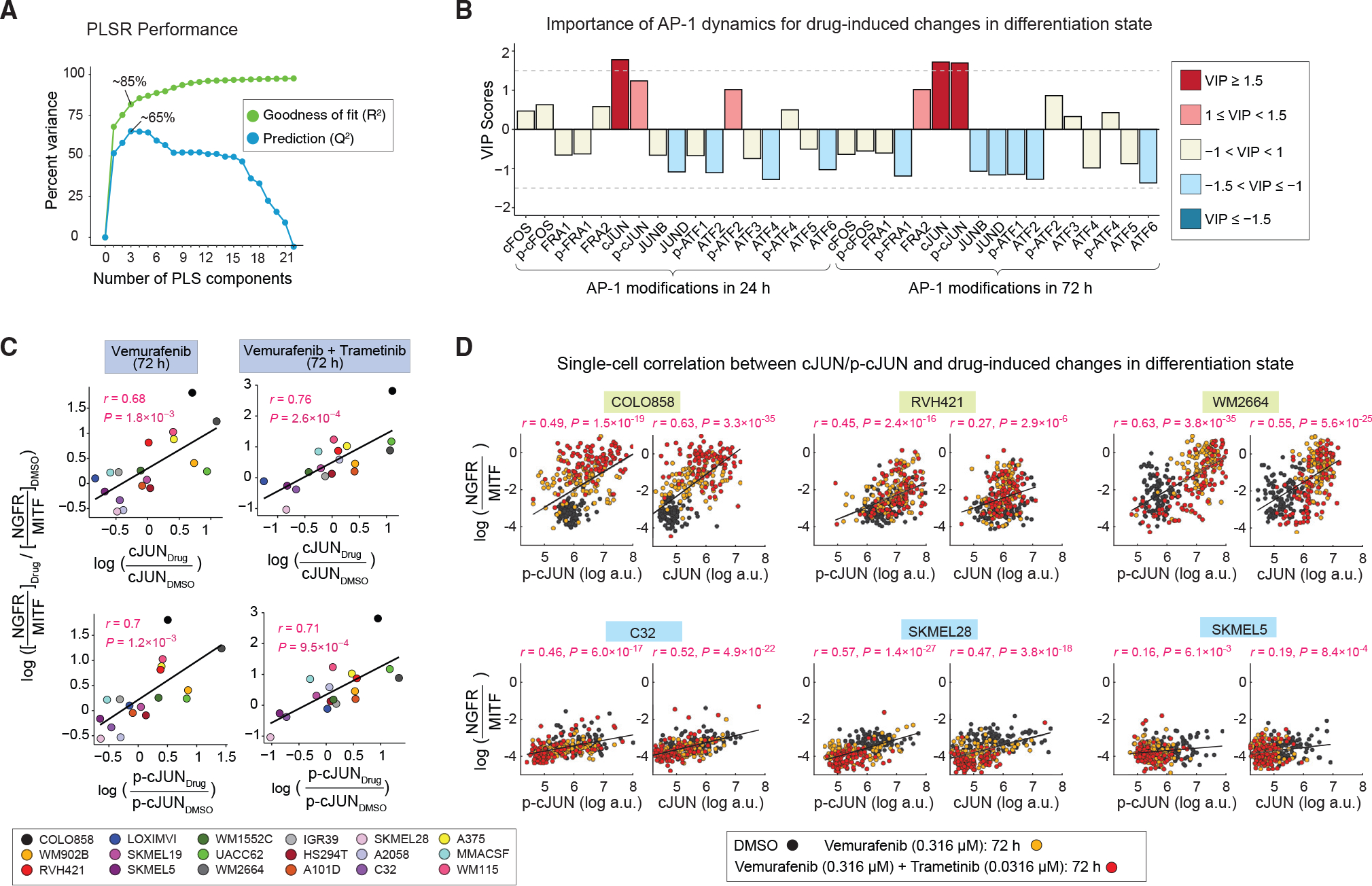
(A) Performance of the PLSR model in predicting drug-induced changes in differentiation states on the basis of drug-induced AP-1 modifications. Model performance was evaluated by computing the fraction of variance in DMSO-normalized differentiation state changes at 72 h explained (R2) or predicted on the basis of leave-one-out cross validation (Q2) with increasing number of PLS components.
(B) PLSR-derived variable importance in projection (VIP) scores, highlighting combinations of DMSO-normalized AP-1 modifications at 24 and 72 h, and their importance for predicting the DMSO-normalized differentiation state changes at 72 h. The sign of the VIP score shows whether the indicated variable (DMSO-normalized AP-1 protein levels at 24 or 72 h) positively or negatively contributes to the response (DMSO-normalized change in differentiation state).
(C) Pearson’s correlation between DMSO-normalized changes in differentiation state and DMSO-normalized cJUN or p-cJUN levels at indicated time points and drug treatment conditions. Each data point represents population-averaged measurements across two replicates for each cell line.
(D) Analysis of covariance between the levels of p-cJUN or c-JUN and drug-induced changes in differentiation state (as evaluated by NGFR-MITF ratio) at the single-cell level across indicated treatment conditions. For each cell line, Pearson’s correlation coefficient was calculated on the basis of 300 randomly sampled cells (100 cells from each treatment condition).
