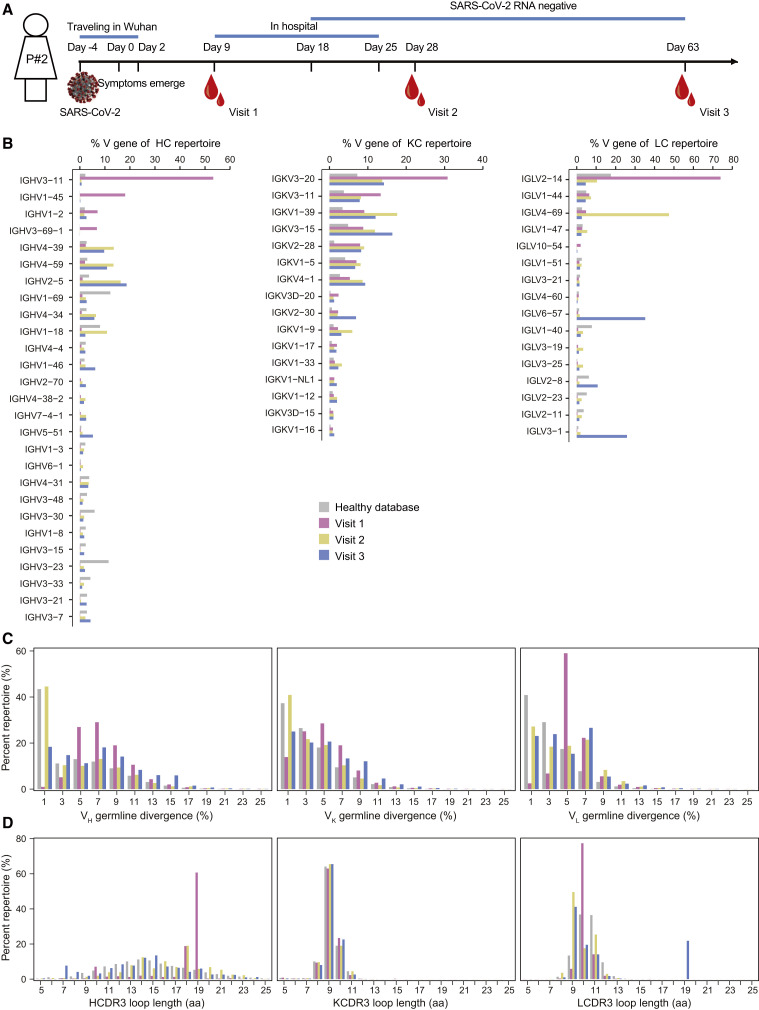
Figure 1.
Antibody repertoire profiles of donor P#2 across the SARS-CoV-2 infection
(A) Schematic representation of experimental design. Blood samples from P#2 were collected at three time points after symptom onset (visit 1: day 9, early infection; visit 2: day 28, clinical recovery; visit 3: day 63, clinical follow-up). The full-length repertoires of variable region of antibodies were obtained by targeted amplification and sequencing on the Illumina MiSeq (2 × 300 bp) platform.
(B–D) Distributions were plotted for (B) germline V gene usage (only those above 1% of the repertoire are shown), (C) degree of SHM, and (D) CDR3 loop length for heavy and light (κ/λ) chains. The antibody database of healthy individuals was downloaded and reanalyzed from the cAb-Rep database (https://cab-rep.c2b2.columbia.edu/). Color coding represents the three time points analyzed, with visit 1 shown in pink, visit 2 in yellow, and visit 3 in blue. The healthy database is shown in gray. HC, heavy chain; KC, κ chain; LC, λ chain.
See also Table S2.