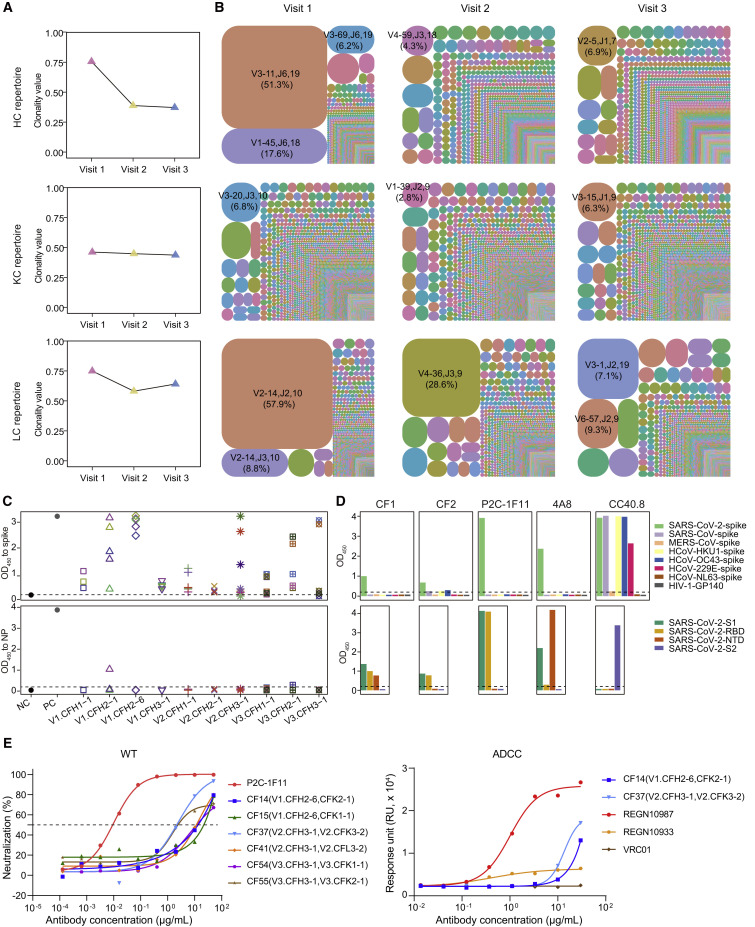
Figure 2.
Clonal expansion of antibody repertoires of donor P#2 at three time points
(A) Diversity of heavy- and light-chain repertoires in different phases. Values were calculated using the Shannon index. HC, heavy chain; KC, κ chain; LC, λ chain.
(B) V-J-CDR3 tree-map of the repertoires. Each rectangle or circle in the tree-map represents a clonotype with the same V and J gene, the same CDR3 length, and more than 80% identity between CDR3 amino acid sequences. The top 10,000 most frequent clonotypes in the antibody repertoires were used for the tree-maps. The size of each rectangle or circle denotes the relative frequency of an individual clonotype. The clonotype ID consists of V gene, J gene, and the CDR3 length, separated by commas. The top clonotypes in each repertoire are listed, and their percentages are indicated in parentheses. The colors of individual clonotypes are randomly chosen, and the colors do not match between plots.
(C) ELISA binding of representative clone-family (CF) antibodies to the SARS-CoV-2 spike (upper panel) and NP (lower panel) proteins. For each time point, selected heavy chains are represented by different shapes, and the selected light chains paired with the heavy chains are represented by different colors. An HIV-1-specific antibody, VRC01, acted as the negative control, while P2C-1F11 and rabbit anti-NP polyclonal antibody were used as positive controls for spike and NP, respectively. NC, negative control; PC, positive control. The OD450 value of ≥0.2 is considered positive.
(D) Binding specificity analysis of representative CF mAbs. CF1 and CF2 were two antibodies from the dominant clonotype in visit 1 repertoires. Three known SARS-CoV-2-spike nAbs were used as control. P2C-1F11 and 4A8, both specific for SARS-CoV-2, are anti-RBD and anti-NTD mAbs, respectively, while CC40.8 targets the S2 subunit of SARS-CoV-2 and cross-reacts with several other human coronavirus spike proteins (Chi et al., 2020; Ju et al., 2020; Zhou et al., 2022). Upper panel: ELISA binding to spike proteins of seven human coronaviruses and HIV-1-GP140 (negative control); lower panel: ELISA binding to different domains of WT SARS-CoV-2. The OD450 value of ≥0.2 is considered positive.
(E) Biological functions of representative CF mAbs. Left panel: Neutralization of six CF mAbs against WT SARS-CoV-2 pseudovirus, with P2C-1F11 as a positive control; Right panel: ADCC of two CF mAbs, with REGN10987, REGN10933, and VRC01 as strong positive control, weak positive control, and negative control, respectively (Hansen et al., 2020). ADCC, antibody-dependent cell-mediated cytotoxicity. The data presented here are means of at least two independent experiments. The curves are representatives of independent experiments with similar results. A cutoff value of 50% is indicated by a horizontal dashed line in neutralization.
See also Figures S2–S4 and Table S3.