FIGURE 4.
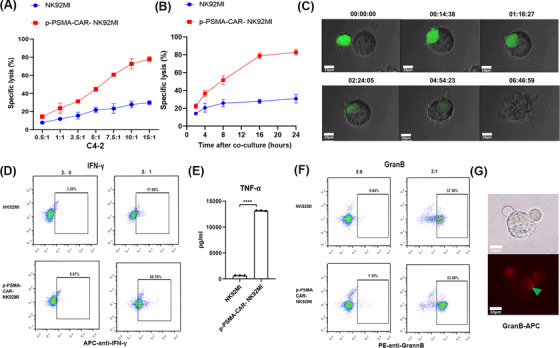
p‐PSMA‐CAR‐NK92MI cells killed a CRPC cell line in vitro. (A) p‐PSMA‐CAR‐NK92MI cells were cocultured with C4‐2 cells at different effector‐to‐target ratios for 4 hours. The specific lysis rate was dose‐dependent. (B) p‐PSMA‐CAR‐NK92MI cells were cocultured with C4‐2 cells at an effector‐to‐target ratio of 1:1 for different time periods. The killing activity of p‐PSMA‐CAR‐NK92MI cells was time‐dependent. (C) The process through which p‐PSMA‐CAR‐NK92MI cells labeled with green fluorescence lysed C4‐2 cells. p‐PSMA‐CAR‐NK92MI cancer cells were edited for green fluorescence. Scale bar: 10 μm. (D) Flow cytometry showed that p‐PSMA‐CAR enhanced IFN‐γ release by NK92MI cells. (E) ELISA revealed that p‐PSMA‐CAR‐NK92MI cells exhibited a significant higher concentration of TNF‐α than NK92MI cells. (F) NK92MI cells engineered with CAR‐CD244‐p‐PSMA secreted more granzyme B than the control after incubation with C4‐2 target cancer cells, as determined by flow cytometry. (G) Immunofluorescence results showed that granzyme B labeled with APC was released into C4‐2 cells. Activated CAR‐NK cells, stimulated by target cell binding, produced granzyme B to lyse target cells. Scale bar: 10 μm. The data are presented as the means ± SEMs from three independent experiments. ****, P <0.0001. Abbreviations: CRPC, castration‐resistant prostate cancer. ELISA, enzyme linked immunosorbent assay; IFN‐γ, interferon gamma; TNF‐α, tumor necrosis factor‐α; APC, allophycocyanin; SEM, standard error of the mean; GranB, granzyme B.
