Gram-negative cell walls are strong enough to withstand ∼3 atm of turgor pressure (40), tough enough to endure extreme temperatures and pHs (e.g., Thiobacillus ferrooxidans grows at a pH of ≈1.5) and elastic enough to be capable of expanding several times their normal surface area (41). Strong, tough, and elastic … the gram-negative cell wall is a remarkable structure which protects the contents of the cell and which has stood the test of time for many, many years. Presumably, these three descriptive traits, have much to do with the tremendous success gram-negative bacteria have had as a life-form on our planet; members of the domain Bacteria inhabit almost all imaginable habitats except those excruciatingly extreme environments in which (some) members of the domain Archaea thrive. Molecular biological methods have not yet given scientists a precise historical record of the origin of gram-negative bacteria, but ancient stromatolites containing fossilized remains of cyanobacterium-like prokaryotes date back to the Archean eon. Over such extraordinary periods of time (much of it when no other life existed), we can imagine that random mutation, selection, and the slowly but ever-changing global environment gave rise to two fundamentally different cell wall formats in Bacteria; gram-positive and gram-negative varieties. Gram-positive cell walls, once thought to be relatively simple structural entities, can be quite different from one another, especially when cell wall turnover is taken into account (8, 9, 25, 29). The cell walls of gram-negative bacteria follow a more general structural format than that of gram-positive bacteria, which is strictly adhered to; gram-negative bacteria have an outer membrane situated above a thin peptidoglycan layer. Sandwiched between the outer membrane and the plasma membrane, a concentrated gel-like matrix (the periplasm) is found in the periplasmic space (7, 9). Because the periplasm exists above the plasma membrane, it is not part of the protoplast, and because the periplasm is differentiated from the external environment by the outer membrane, it is not part of the “outside.” It is in fact an integral compartment of the gram-negative cell wall (5). Together the plasma membrane and the cell wall (outer membrane, peptidoglycan layer, and periplasm) constitute the gram-negative envelope (5, 9).
Our entire perception of gram-positive and gram-negative walls ultimately relies on the response of bacteria to Gram staining. Unwittingly, in 1884, Christian Gram developed a staining regimen for light microscopy which differentiated between these two types of bacteria because of the chemical composition and structural format of their cell walls. Because gram-negative bacteria possess a lipid-rich outer membrane (as well as a plasma membrane) and a thin peptidoglycan layer, the alcohol decolorizing step of Gram staining washes the primary stain (crystal violet) from the cells and the secondary stain (carbol fuchsin or saffranin) colors the bacteria red (57). Gram-positive bacteria are enshrouded in thicker, more resilient cell walls which do not allow the crystal violet to be removed and, accordingly, remain purple (57). Although the vast majority of bacteria adhere to the color differentiation of the Gram stain, to the chagrin of microbiological taxonomists, some bacteria refuse to obey; these are called gram-variable bacteria (6). Members of the Archaea cannot be easily differentiated by Gram staining (10). Interestingly, the staining response of gram-variable bacteria and archaea is also due to their cell wall composition and structure (6, 10).
Advances in identifying gram-negative cell wall components, their cytoplasmic synthetic and plasma membrane translocation routes, and their individual functional attributes have been electrifying over the last decade. This is primarily due to their intricate dissection by modern molecular techniques. There are several up-to-date reviews describing specific gram-negative cell wall systems which emphasize their molecular biological aspects (28, 56, 60), and I will not revisit them in this minireview.
OUTER MEMBRANE
Although layers that are more external (such as capsules, S-layers, and sheaths) (5) can reside above the outer membrane, this lipid-protein bilayer is usually considered to be the outermost layer of the gram-negative wall. It is a membrane which possesses proteins, phospholipids, and lipopolysaccharides (LPSs) and which separates the external environment from the periplasm. Because bacteria rely on diffusion for nutrition and the dissemination of wastes, the outer membrane must be porous to certain substances (hence the addition of porins which assemble into pores) (e.g., 15) and must be capable of transporting others (e.g., iron). Even hydrophobic compounds can also make their way through (54). Yet, the outer membrane cannot be too porous, since larger periplasmic constituents which are vital to the cell’s livelihood must be retained. For example, the periplasm contains binding proteins for amino acids, sugars, vitamins, and ions, as well as degradative and detoxifying enzymes. It can also act as a reservoir for such surface-associated components as certain pilins, S-layer proteins, and virulence factors (e.g., proelastin of Pseudomonas aeruginosa) (19, 39). Because of the concentration of these constituents and so-called membrane-derived oligosaccharides (38), the osmolarity of the periplasm may approach the osmolarity of the cytoplasm. Koch recently argued that an osmotic gradient does not exist over the outer membrane (40).
One of the few ways in which researchers can image the outer membrane has been with a transmission electron microscope (TEM). This membrane has been difficult to preserve for TEM observation. Although some of the outer membrane proteins (OMPs) can be associated with the underlying peptidoglycan layer (e.g., Braun’s lipoprotein in Escherichia coli), many of the bilayer constituents (especially the lipids) are fluid and in continual rapid motion. This is often a difficult concept for researchers to understand; at the nanoscale level, components such as LPS are constantly in motion. They move at high speeds laterally around the cell, and they are, at the same time, rotating on their long axis. Even the O-side chains are flexing back and forth and seem to be driven entirely by entropy (53). Environmental conditions (e.g., temperature) and molecular associations (e.g., LPS-OMP interactions) affect free motion.
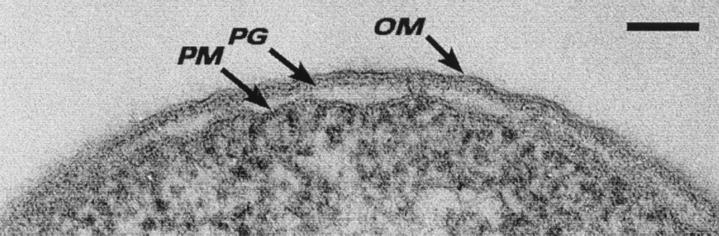
For electron microscopy, researchers have to (somehow) instantaneously stop the movement of molecules in the membrane and denature any degradative enzymes which could affect bilayer structure. At the same time, underlying supportive structures for the membrane (such as the periplasm and peptidoglycan layer) must be accurately maintained and preserved. Traditionally, a procedure using conventional embedding and thin sectioning has been considered to be one of the best ways to examine the outer membrane. For this, extremely toxic chemical fixatives (such as glutaraldehyde and osmium tetroxide) are used to increase the covalent bonding in the specimen so that it can withstand organic solvent dehydrating and plastic embedding protocols. Fixation, dehydration, and embedding rely on the diffusion of the chemical agents into the specimen (often at room temperature), and the process takes hours. Delicate structures, such as membranes, are notoriously poorly preserved, the periplasm is extracted, and the outer membrane usually shows an artificial wavy configuration (Fig. 1).
FIG. 1.
Thin section of the cell envelope of E. coli K-12 after conventional embedding. The periplasmic space is empty of substance, and the peptidoglycan layer (PG), outer membrane (OM), and plasma membrane (PM) can be seen. Bar = 100 nm.
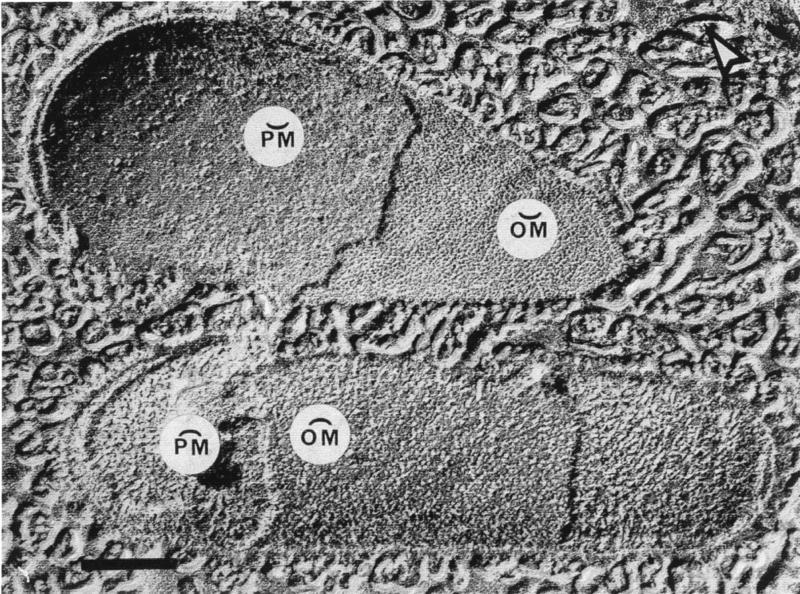
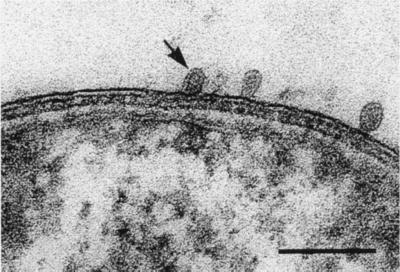
With the recognition that extremely rapid freezing of biological samples could physically fix their structure, almost instantaneously, the technique of freeze-etching began to be used in microbiology. In this technique, bacteria are frozen so rapidly that vitreous (or amorphous) ice forms and molecular motion stops. The ice-cell matrix is a hard solid and can be fractured in a freeze-etching device. Since the fracture plane follows the regions of least bond energy within a cell, membranes are most frequently cleaved through their hydrophobic domains and intrinsic membrane proteins are exposed. Fortuitous fractions through gram-negative bacteria can reveal the inner and outer faces of membranes by exposing concave and convex surfaces (Fig. 2). Freeze-etching of a large variety of gram-negative bacteria has consistently shown that the outer membrane is strongly bonded together, and for this reason, fractions through its hydrophobic domain are rare; the preferred fracture plane is through the plasma membrane (Fig. 2).
FIG. 2.
Freeze-etching of two E. coli K-12 cells in which the fracture planes have travelled through the cell envelope. The upper cell shows concave fractures through the outer membrane (OM) and plasma membrane (PM), whereas the lower cell shows convex fractures of the same membranes. The particles (or holes) in these membrane fractures correspond to intramembranous protein complexes. The arrowhead in the upper right-hand corner of the image points out the shadow direction. Bar = 500 nm. (Reprinted from reference 9.)
The newer cryotechnique of freeze-substitution has been more informative. Here, cells are rapidly frozen (as with freeze-etching) and immersed in a cryosubstitution mixture of osmium tetroxide and uranyl acetate in anhydrous acetone, and the mixture was held at −80°C for 48 h (26). The bacteria are chemically fixed, stained, and dehydrated while being maintained in an ultrafrozen state. Eventually, they can be embedded in plastic at room temperature and thin sectioned. Kazunobu Amako’s group in Japan was the first to apply freeze-substitution to a gram-negative bacterium (1) and produced remarkable images of E. coli that made all microscopists who study microorganisms reevaluate their previous work. This was soon followed by the remarkable images of E. coli’s cell envelope from Eduard Kellenberger’s group in Switzerland for which the term periplasmic gel was coined (30). Here, for the first time, was tangible microscopic proof that a rich, dense periplasm existed in the periplasmic space of a gram-negative cell.
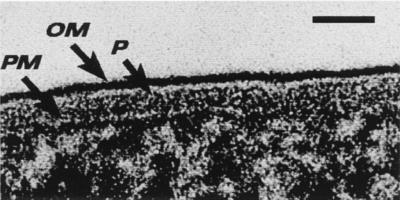
One of the unusual features of the outer membrane is its asymmetric distribution of lipids over its inner and outer faces. The outer face contains (virtually) all of the LPS, whereas the inner face has most of the phospholipid. LPS contains more charge per unit of surface area than any phospholipid, and most of this charge is anionic at neutral pH because of exposed phosphoryl and carboxyl groups which can be readily ionized. The outer face of the outer membrane is highly charged and is highly interactive with cations in the outside milieu, so interactive, in fact, that these anionic groups can be sites for the development of fine-grained minerals in natural environments (22). This reactivity of LPS is also convenient for the study of the outer membrane by TEM since the outer face reacts strongly with heavy metal stains if the lipid asymmetry is retained. Freeze-substitution preserves this asymmetry (Fig. 3) and reveals the length and distribution of the LPS O-side chains (Fig. 4). It is not uncommon to find more than one LPS species on an outer membrane at a time. P. aeruginosa PAO1 has two LPS moieties; A-band LPS (the common antigen) which is a short chain and neutral in charge, and B-band LPS (serotype LPS) which is a longer chain and (overall) electronegative. Freeze-substitution has shown that the O-side chains of B-band LPS can extend up to 40 nm from the outer membrane (Fig. 4) (43). Images such as these have suggested that O-side chains are frequently in their extended conformation (43).
FIG. 3.
Freeze-substitution image of the E. coli K-12 cell envelope for direct comparison with Fig. 1. The periplasmic space is filled with periplasm (P) (the so-called periplasmic gel), and the peptidoglycan layer is not visible. The outer face of the outer membrane (OM) is densely stained, because the LPS retains its asymmetric location in this region of the bilayer and is more highly charged than the phospholipid on the inner face of the OM. The periplasm is bounded by the OM and the plasma membrane (PM). Bar = 25 nm. (Reprinted from reference 9.)
FIG. 4.
Freeze-substitution image of the P. aeruginosa PAO1 cell envelope showing the long O-side chains of the B-band LPS extending ∼40 nm from the face of the outer membrane (arrow). Bar = 35 nm. (Reprinted from reference 9.)
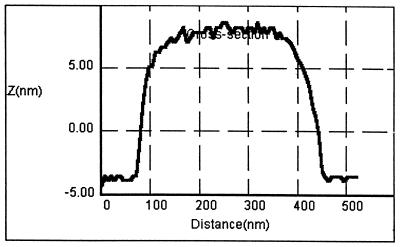
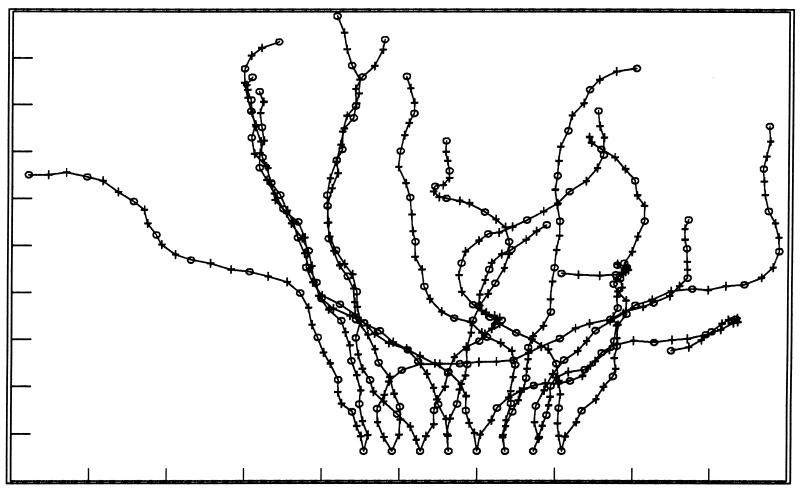
However, recent atomic force microscopy (AFM) measurements have not verified this observation (64), which presents an enigma. In these experiments, AFM was used as a pressure-measuring technique where the AFM tip is lowered down to the level where it can first monitor pressure resistance from the surface molecules. For isolated B-band LPS, resistance cannot be detected until the AFM tip is ∼10 nm from the surface of a LPS micelle (Fig. 5). This then begs the question … how can a cryoelectron microscopical technique show B-band O-side chains extending up to 40 nm away from the outer membrane’s bilayer structure and AFM pressure measurements detect the O-side chains only ∼10 nm away? To solve this discrepancy, several B-band LPS molecules have been dynamically modeled together using brush theory as they interact with one another on the outer membrane’s surface. Remarkably, this analysis has shown the O-side chains to be in constant motion; the side chains are flexing and rotating continuously so that only a proportion of the chains are in their ∼40-nm-extended configuration at a single point in time (Fig. 6). These are the side chains that are captured in a single instant by the rapid-freezing, freeze-substitution technique (Fig. 4). Because longer times are required to perform an AFM pressure measurement and because the O-side chains are so rapidly moving, the pressure measurements do not detect the flexible O-side chains but detect only the core oligosaccharide molecules attached to lipid A which have more restricted movement.
FIG. 5.
AFM tracing over a single hot-phenol-extracted A- and B-band LPS micelle from P. aeruginosa PAO1 obtained under water (pH 7.0) after the micelle had attached to a silicon nitride surface. The 12-nm height (Z on the y axis) of the vesicle represents both the lower and upper faces of the LPS bilayer and is consistent with only the lipid A plus the core oligosaccharide being detected on the 300-nm-diameter micelle. Yet, sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the preparation showed that the O-side chains of both LPSs are present, and freeze-substitution images of intact outer membrane surfaces (Fig. 4) revealed that the B-band O-side chain extends ∼40 nm. We believe the O-side chains are so rapidly moving under the aqueous conditions of our AFM experiment that the AFM tip cannot detect them.
FIG. 6.
Schematic diagram showing the different alignments (i.e., movement) of the B-band O-side chains of P. aeruginosa as we perceive them from the AFM experiments shown in Fig. 5 (53). The model assumes that all ionizable groups are charged. The core oligosaccharide and lipid A moieties are not shown but would be at the bottom of the diagram.
This rapid motion of O-side chains has important consequences. The B-band side chains are charged with an overall electronegative (anionic) charge at neutral pH. Intuitively, the chains should (then) interact with environmental electrolytes such as multivalent metal ions. They should be salt bridged by ions, such as Ca2+, Cu2+, Mg2+, or Fe3+, and be relatively immobile. This does not seem to be the case (44). It is possible that the side chains are moving so rapidly that metal ion salt bridges are not energetically possible. If this were true, then the same binding constraints should apply to other ionizable external components such as immunoglobulins or even components on bacteriophage tails or phagocytotic cells (i.e., receptors). Certainly, these components do bind to bacterial surfaces but maybe the length of LPS O-side chains and their rapid motion could, in a general way, affect how strongly such external components can interact.
PERIPLASM AND MEMBRANE VESICLES (MVS)
Cryo-TEM has not helped in the elucidation of the peptidoglycan layer (or murein sacculus) of gram-negative bacteria. This is not so much because the layer is not preserved as well as usual but because it is difficult to see it in thin sections of freeze-substituted cells. Cryopreservation has retained so much substance in the periplasmic space (where the peptidoglycan layer resides) that the layer is obscured (Fig. 3) (1, 9, 27, 30). For the first time, researchers can actually see how much material resides in the periplasmic space, which indirectly points out the importance of this region to the vitality of gram-negative cells. Periplasmic enzymes, trafficking proteins, secreted materials, and newly synthesized outer membrane- or peptidoglycan layer-directed components all reside in this space at some point during the bacterium’s metabolic life (7). It is possible that membrane junctions occur between outer and plasma membranes (the so-called Bayer’s junctions) to aid in the transfer of outgoing materials (3), but the peptidoglycan layer still separates the two membranes at these junctions. Freeze-substitution, though, has certainly revealed so much substance within the periplasm that it should behave as a gel (30).
Periplasm.
It is important to recognize the full scope of the variety of macromolecules that can inhabit the periplasm of a gram-negative bacterium. It is clear that a bacterium relies on environmental signals to influence its synthesis and secretion pathways. Quorum sensing can help coordinate such a response in relatively large bacterial populations or even in biofilms (13, 16, 52). For example, many of the virulence factors of P. aeruginosa, which is an opportunistic pathogen often implicated in cystic fibrosis, require induction before they are secreted (51). In fact, even osmotic differences between the cytoplasm and periplasm can trigger the production of membrane-derived oligosaccharides (38). The periplasm, then, is a region between the two bilayered membranes of the gram-negative cell envelope which is in dynamic flux possessing an ever-changing variety of macromolecules which reflect the cell’s metabolic and environmental status.
MVs.
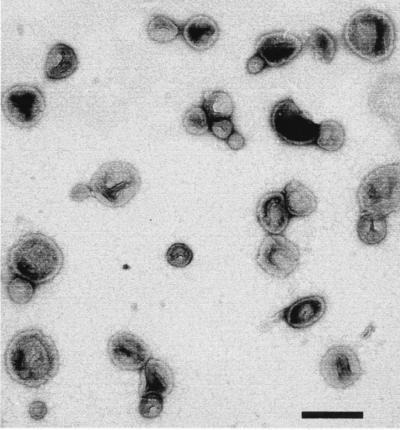
Gram-negative cell walls have a dynamic feature that is not seen in their gram-positive counterparts … outer membrane vesicles are constantly being discharged from the surface of the cell during bacterial growth. As the vesicles are being extruded from the surface, they entrap some of the underlying periplasm so that they are actually small particles of gram-negative cell wall. They possess OMPs, LPS, phospholipids, and periplasmic constituents, all situated as they normally would be in a bacterium, but on a much smaller scale. These 50- to 250-nm-diameter spherical, bilayered, membranous structures (Fig. 7) are released from the surfaces of virtually all gram-negative bacteria (11, 34, 47). Reports of these vesicles date back ∼30 years (47), and additional reports have been published since then (17, 18, 21, 32, 42, 45, 59, 61, 62, 65). The general importance of their release has recently been recognized, and they are called membrane vesicles or MVs (11, 32, 34). MVs are found emanating from gram-negative bacteria growing in the planktonic or biofilm mode (12), on solid or liquid media (34), as swarming cultures (23), and in natural environments (Fig. 8) (12). The MVs from P. aeruginosa have been the most intensively studied (30–32, 45).
FIG. 7.
Negative-stained n-MVs which have been isolated and purified from P. aeruginosa as previously described (32). Bar = 250 nm.
FIG. 8.
Thin section of an unidentified gram-negative bacterium found in a freshwater biofilm in a river near laboratory. This bacterium possesses a microcapsule and is liberating a prodigious amount of MVs. Bar = 1 μm.
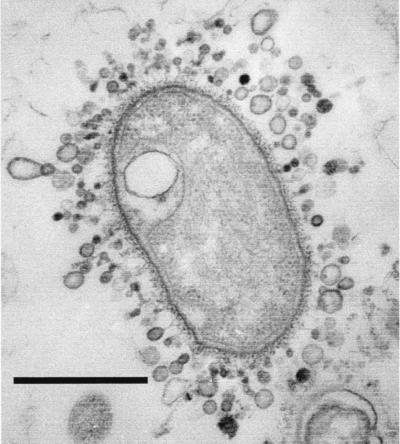
As MVs form blebs from the outer membrane, they encapsulate periplasm so that once they are free of the bacterium, the MV becomes a small membrane-bound vessel of periplasmic constituents (Fig. 9). This phenomenon was aptly demonstrated for P. aeruginosa MVs, since they were found to contain protease, phospholipase C, alkaline phosphatase, and an autolysin (32, 33, 45). These MVs also contained proelastase which was proof of periplasm encapsulation, since the pro amino acid sequence is cleaved from the enzyme only after translocation across the outer membrane during its normal secretion (19, 39). It is important to recognize that the method by which MVs are released from gram-negative cell surfaces ensures that the natural outer membrane arrangement is generally maintained. The normal low curvature of the outer membrane (while it resides on the bacterium) is abruptly changed to the high-curvature form of the vesicle (Fig. 9), but LPS, phospholipids, and OMPs are still integral constituents of the MV’s bilayer, and the outer membrane LPS-phospholipid asymmetry is retained (32). Interestingly, there is one important alteration from normal outer membranes in the P. aeruginosa system. Instead of possessing both A- and B-band LPS (as the outer membrane does), only B-band (or serotype-specific) LPS is present in MVs (32). Somehow, P. aeruginosa, by pooling B-band LPS into small, localized regions on its outer membrane, forces the membrane of these regions into high-curvature structures, thereby encouraging the formation of blebs and release of MVs (32).
FIG. 9.
Thin section of P. aeruginosa PAO1 showing the development of n-MVs before they are liberated from the cell. The arrow points to one vesicle in which the membrane bilayer and the periplasm within its lumen (i.e., electron-dense area inside the vesicle) can be seen. Bar = 250 nm.
Peptidoglycan hydrolases of MVs.
Gram-negative bacterial periplasm normally contains autolysins which along with penicillin-binding proteins are used to help fabricate the peptidoglycan layer as a bacterium grows. Because of this, it is possible that MVs entrap a proportion of these peptidoglycan-hydrolyzing enzymes during MV formation.
P. aeruginosa produces 11 autolysins which can be renatured after hot sodium dodecyl sulfate extraction (4). One of these autolysins, the major 26-kDa peptidoglycan hydrolase (PGase) is packaged into MVs (45). There seemed to be little point for this PGase encapsulation until experiments were devised to see whether the MVs could affect the integrity of other bacteria. Remarkably, MVs can lyse other bacteria as long as these bacteria are suffering from low nutrition and growing poorly (46). MVs can attack both gram-positive and -negative bacteria, and the potency of MV attack depends on the peptidoglycan chemotype of the attacked cells; those with chemotypes identical or similar to the chemotype of P. aeruginosa (A1γ) were readily lysed. A closer inspection by electron microscopy revealed that MVs attack gram-positive and -negative surfaces in a different manner (33). For gram-positive bacteria, MVs adhere to the surface of the cell wall, break open, and hydrolyze the peptidoglycan immediately under the adherence junction. The same mechanism occurs even if the cell wall has a S-layer on top of it (36); presumably, after the MV has broken open, the 26-kDa PGase can penetrate the lattice network of the S-layer and attack the underlying cell wall. In this way, whether or not gram-positive cells possess a S-layer, MVs attack their surfaces through a single-hit route which produces a single large lesion in the cell wall (33).
MVs attack gram-negative bacteria in a much different manner. Here, MVs adhere to the outer membrane and rapidly fuse into it (33). In so doing, the luminal contents (including the PGase) are released directly into the periplasmic space of the recipient cell. Once inside the periplasmic space, the PGase can fully diffuse around the protoplast and hydrolyze the peptidoglycan layer at a number of different sites so that multisite lysis can occur.
MV-mediated lysis of bacteria does not readily occur if the recipient cells are actively growing and dividing; it occurs only when they are under the constraints of poor nutrition. Presumably, with active growth, the MV-induced cell wall lesions can be repaired by the recipient’s normal cell wall turnover cycle, and as the cell grows and divides, the MV PGase is gradually diluted. Because poor growth conditions are required for efficient killing of other bacteria by MVs, we have speculated that MVs are a predatory response by the donor cell (in this case, P. aeruginosa) to increase its nutrient load under poor environmental conditions (33). The MVs lyse surrounding foreign bacteria, thereby liberating highly complex nutritious constituents for the use of donor cells. The idea of foreignness is important in this concept; MVs would not kill neighboring cells of an identical strain (i.e., those surrounding the donor) because the MV PGase would be one of their own normal autolysins and could be regulated as such. In this way, MVs would benefit the entire population of a single or identical clone in a natural environment. Since MVs are frequently encountered in biofilms (12), this could be one way in which microcolonies retain their integrity and sustenance at the expense of the surrounding microflora.
MVs and virulence.
The pathogenicity of a variety of gram-negative bacterial pathogens relies, at least in part, on their ability to secrete a number of virulence factors into the menstruum surrounding their targeted tissue (e.g., hemolysin, aerolysin, verotoxin, etc.). These factors diffuse into the tissue and begin to break it down. Yet, free diffusion of such metabolically expensive macromolecules can be wasteful. Once free of the pathogen, the factors are diluted by diffusion, and external host constituents (e.g., hydrolytic enzymes, antibodies, and other serum constituents) can inactivate them.
MVs could provide an alternative route for the delivery of virulence factors. For example, the MVs of P. aeruginosa, Proteus mirabilis, and Serratia marcescens package phospholipase C, proteases, proelastase, and hemolysins (23, 24, 32). It is probable that MVs from Borrelia, enterotoxigenic E. coli, Haemophilus, Neisseria, and Vibrio species also contribute to the virulence of these other pathogens (17, 42, 59, 61, 62). The constituents which are packaged into the lumens of MVs are protected from the action of inactivating environmental enzymes by the MV’s bilayer which contains LPS, phospholipids, and OMPs. Because LPS is endotoxic and OMPs can be highly antigenic, MVs can be even more antagonistic to the host.
Recently, it has been discovered that a chromosome-encoded β-lactamase in P. aeruginosa (an enzyme which normally hydrolyzes β-lactam antibiotics in the periplasm of these cells) can also be packaged into MVs (14). This raises the intriguing possibility that β-lactamase-containing MVs could be discharged from pathogens at their infection sites in tissue to increase the breakdown of β-lactam antibiotics in the local tissue environment. Because MVs contain porins as part of their OMP complement, the antibiotic readily diffuses into these MVs and is inactivated. This could be a general strategy for reducing antibiotic concentrations at infection sites, and it could also be one of the ways in which chronic, hard-to-treat infections (such as cystic fibrosis) are maintained by the infecting bacterium (14). Since MVs can readily fuse to the outer membranes of a wide range of other gram-negative bacterial pathogens (37), thereby emptying their luminal contents into the periplasm of recipient cells, β-lactamase (and other antibiotic-inactivating enzymes) could be physically transferred from a donor cell to a non-β-lactamase-producing recipient (e.g., following the cystic fibrosis analogy, from P. aeruginosa [donor] to Burkholderia cepacia [recipient]). In this way, a varied group of gram-negative nonproducers in close proximity to a donor cell could withstand antibiotic treatment and contribute to the infection without these same bacteria having the genetic capacity to produce the inactivating enzyme themselves. This type of synergy between pathogens has not been suggested before and certainly requires further investigation. Interestingly, a study by Dorward et al. (18) has shown that MV-mediated transfer of an antibiotic resistance plasmid can occur between two strains of Neisseria gonorrhoeae. It is therefore possible that MVs influence antibiotic resistance in other bacteria in two ways: the physical dissemination of preformed antibiotic-inactivating enzymes into their periplasm and the delivery of antibiotic resistance plasmids to nonproducing strains.
Possible medical application of MVs.
Early in our research with P. aeruginosa MVs, we learned that membrane surface-active agents such as gentamicin could increase the production of MVs about threefold (32). In so doing, small amounts of the aminoglycoside became entrapped in the MVs. Gentamicin-containing MVs (g-MVs) also differ from natural MVs (n-MVs) by containing small amounts of A-band LPS in addition to B-band LPS. g-MVs have a greater size variability than n-MVs, and they sometimes contain plasma membrane and small amounts of DNA (32). Yet, the increased production of MVs and their retention of gentamicin (∼5 to 10 ng of gentamicin per μg of MV protein) make g-MVs a tempting vehicle for antibiotic delivery to hard-to-kill pathogens. For example, g-MVs can kill Pseudomonas spp. which are normally impermeable to aminoglycoside antibiotics (33). This is because g-MVs fuse to the normally impermeable outer membrane of the resistant strain and deliver gentamicin into the periplasm where it can be actively imported to the cytoplasm and inhibit protein synthesis. Although g-MVs possess small amounts of gentamicin, the antibiotic is targeted to the correct region of the cell for strong inhibitory effect.
Aminoglycoside antibiotics also have trouble entering eukaryotic tissue to inhibit the growth of intracellular bacterial pathogens. Recent experimentation with Shigella flexneri g-MVs (these contain 85 ng of gentamicin per μg of MV protein [35]) has revealed that these particular MVs contain the invasion proteins (49, 50) of this intracellular pathogen. For this reason, when epithelial cell lines are treated with S. flexneri g-MVs, the vesicles are engulfed by the tissue cells and the gentamicin is eventually found in the eukaryotic cytoplasm (35). Indeed, when this same tissue was first infected with S. flexneri and then treated with S. flexneri g-MVs, a substantial quantity (∼1.5 log10 CFU) of the intracellular pathogen was cleared from the tissue within 60 min because the g-MVs conveyed gentamicin to the cells (35).
These experiments suggest that MVs could be used to deliver drugs to a number of different prokaryotic and eukaryotic systems.
Another possible medical application for MVs (in this case, n-MVs) is as new vaccine agents, because when MVs are released from the bacterium, they contain the surface identity of the donor bacterium. The LPS is serotype LPS and is strain specific, and the same is true for the OMPs. Important, highly antigenic virulence factors are also present. Strains of P. mirabilis and S. marcescens which possess adhesins (pili or fimbriae) also have these structures associated with the MVs (23, 24). Clearly, MVs are strongly antigenic particulate structures which could also possess natural adjuvant qualities for enhancing an immune response.
A major obstacle in the use of MVs as particulate vaccines is the presence of endotoxic LPS; if this substance cannot be detoxified, the use of MVs as vaccine agents would be limited to oral route administration. Since detoxification requires chemical manipulation of the lipid A of LPS and, possibly, reformulation of MV composition and structure, it may be more promising to look more closely at administering MVs via the oral route. MVs from a number of gram-negative pathogens can be integrated into the outer membranes of members of the family Enterobacteriaceae (37), and it may be possible to use oral, attenuated vaccine strains containing incorporated foreign MVs as multiepitope vaccine candidates.
Are MVs a new form of secretion?
Over the last 2 decades, a number of different secretion systems have been elucidated (55). Most systems involve steps necessary to translocate a preformed polypeptide across the plasma membrane. During this translocation, the polypeptide can be matured (e.g., a signal sequence can be cleaved off or disulfur bonds can be formed from thiols) (2, 20, 31, 55) so that the periplasmic form is different from the originally synthesized form. In addition, some periplasmic polypeptides must undergo further transformation before they are expelled into the external milieu (e.g., proelastase is cleaved as it passes through the outer membrane to become active elastase) (19, 39), or two (or more) periplasmic components can anneal together only after they have been expelled (e.g., subunits A and B of cholera toxin [48]). Only in rare instances are the polypeptides actively translocated through the outer membrane (58, 63). Usually, simple diffusion through outer membrane pores is invoked for the vast number of these relatively large compounds, sometimes with the assistance of outer membrane-plasma membrane adhesion zones (3). Certainly they do make it through this last limiting membrane, and soluble secretion products can be readily found in the external fluid.
Are these the only routes for the final secretion steps of such compounds? Maybe not. MVs appear to be an alternative route which, so far, has been poorly recognized by those in the secretion field. Yet, the evidence is undeniable … MVs are produced by virtually all gram-negative bacteria, they are commonly filled with components considered to be secretion products, and they have the capacity to protect these products by their limiting membrane as they are conveyed to other cells, whether these cells be prokaryotic or eukaryotic.
Are MVs a new form of secretion? Intuitively, MVs are not a brand-new structural device for gram-negative bacteria. Because of their complexity, they must have slowly evolved into the systems we see today … we have just not recognized their importance until recently!
ACKNOWLEDGMENTS
Some of the research reviewed here comes from the work of students, postdoctoral fellows, and scientists who have passed through my laboratory over the last few years. They will know who they are, and I thank them all. The biophysical aspects of LPS were done in collaboration with M. Jericho, Dalhousie University, and D. Pink, St. Francis Xavier University.
My research has been supported by grants from the Natural Sciences and Engineering Council (both Research and Major Facilities Access grants) and from the Canadian Bacterial Disease Network which is a National Centre of Excellence.
REFERENCES
- 1.Amako K, Murata K, Umeda A. Structure of the envelope of Escherichia coli observed by the rapid freezing and substitution fixation method. Microbiol Immunol. 1983;27:95–99. doi: 10.1111/j.1348-0421.1983.tb03571.x. [DOI] [PubMed] [Google Scholar]
- 2.Åslund F, Beckwith J. The thioredoxin superfamily: redundancy, specificity, and gray-area genomics. J Bacteriol. 1999;181:1375–1379. doi: 10.1128/jb.181.5.1375-1379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer M E. Zones of membrane adhesion in the cryofixed envelope of Escherichia coli. J Struct Biol. 1991;197:268–280. doi: 10.1016/1047-8477(91)90052-x. [DOI] [PubMed] [Google Scholar]
- 4.Bernadsky G, Beveridge T J, Clarke A J. Analysis of the sodium dodecyl sulfate-stable peptidoglycan autolysins of select gram-negative pathogens by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1994;176:5225–5232. doi: 10.1128/jb.176.17.5225-5232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beveridge T J. Ultrastructure, chemistry, and function of the bacterial cell wall. Int Rev Cytol. 1981;72:229–317. doi: 10.1016/s0074-7696(08)61198-5. [DOI] [PubMed] [Google Scholar]
- 6.Beveridge T J. Mechanism of gram variability in select bacteria. J Bacteriol. 1990;172:1609–1620. doi: 10.1128/jb.172.3.1609-1620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beveridge T J. The periplasmic space and the concept of the periplasm in gram-positive and gram-negative bacteria. ASM News. 1995;61:125–130. [Google Scholar]
- 8.Beveridge T J. The ultrastructure of gram-positive cell walls. In: Fischetti V, Novick R, Ferretti J, Portnoy D, Rood J, editors. Gram-positive pathogens. Washington, D.C: American Society for Microbiology; 1999. pp. 3–10. [Google Scholar]
- 9.Beveridge T J, Graham L L. Surface layers of bacteria. Microbiol Rev. 1991;55:684–705. doi: 10.1128/mr.55.4.684-705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beveridge T J, Schultze-Lam S. The response of selected members of the Archaea to the Gram stain. Microbiology. 1997;142:2887–2895. doi: 10.1099/13500872-142-10-2887. [DOI] [PubMed] [Google Scholar]
- 11.Beveridge T J, Kadurugamuwa J L. Periplasm, periplasmic spaces, and their relation to bacterial wall structure: novel secretion of selected periplasmic proteins from Pseudomonas aeruginosa. Microb Drug Resist. 1996;2:1–8. doi: 10.1089/mdr.1996.2.1. [DOI] [PubMed] [Google Scholar]
- 12.Beveridge T J, Makin S A, Kadurugamuwa J L, Li Z. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20:291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 13.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 14.Ciofu, O., T. J. Beveridge, J. Kadurugamuwa, J. Walther-Rosmussen, and N. Høiby. Chromosomal β-lactamase is packaged into membrane vesicles from Pseudomonas aeruginosa. Submitted for publication. [DOI] [PubMed]
- 15.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 16.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 17.Devoe I W, Gilchrist J E. Research of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J Exp Med. 1973;138:1156–1166. doi: 10.1084/jem.138.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorward D E, Garon C F, Judd R C. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fecycz I T, Campbell J N. Mechanism of activation and secretion of a cell-associated precursor of an exocellular protease of Pseudomonas aeruginosa 34362A. Eur J Biochem. 1985;146:35–42. doi: 10.1111/j.1432-1033.1985.tb08616.x. [DOI] [PubMed] [Google Scholar]
- 20.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system in Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 21.Forsberg C W, Beveridge T J, Hellstrom A. Cellulase and xylanase release from Bacteroides succinogenes and its importance in the rumen environment. Appl Environ Microbiol. 1981;42:886–896. doi: 10.1128/aem.42.5.886-896.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortin D, Ferris F G, Beveridge T J. Surface-mediated mineral development by bacteria. Rev Mineral. 1997;35:161–180. [Google Scholar]
- 23.Fu, H., and T. J. Beveridge. 1999. Unpublished data.
- 24.Fu, H., N. Allen, and T. J. Beveridge. 1999. Unpublished data.
- 25.Giesbrecht P, Kersten T, Maidhof H, Werke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham L L, Beveridge T J. Evaluation of freeze-substitution and conventional embedding protocols for routine electron microscopic processing of eubacteria. J Bacteriol. 1990;172:2141–2149. doi: 10.1128/jb.172.4.2141-2149.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham L L, Harris R, Villiger W, Beveridge T J. Freeze-substitution of gram-negative eubacteria: general cell morphology and envelope profiles. J Bacteriol. 1991;172:1623–1633. doi: 10.1128/jb.173.5.1623-1633.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinrichs D E, Yethon J A, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 29.Higgins M L, Shockman G D. Study of a cycle of cell wall assembly in Streptococcus faecalis by three-dimensional reconstruction of thin sections of the cell. J Bacteriol. 1976;137:1346–1358. doi: 10.1128/jb.127.3.1346-1358.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hobot J A, Carlemalm E, Villiger W, Kellenberger E. Periplasmic gel: new concept resulting from the reinvestigation of bacterial cell envelope ultrastructure by new methods. J Bacteriol. 1984;160:143–152. doi: 10.1128/jb.160.1.143-152.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadurugamuwa J L, Beveridge T J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadurugamuwa J L, Beveridge T J. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadurugamuwa J L, Beveridge T J. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J Antimicrob Chemother. 1997;40:615–621. doi: 10.1093/jac/40.5.615. [DOI] [PubMed] [Google Scholar]
- 35.Kadurugamuwa J L, Beveridge T J. Delivery of the non-membrane-permeative antibiotic gentamicin into mammalian cells using Shigella flexneri membrane vesicles. Antimicrob Agents Chemother. 1998;42:1476–1483. doi: 10.1128/aac.42.6.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadurugamuwa J L, Mayer A, Messner P, Sára M, Sleytr U B, Beveridge T J. S-layered Aneurinibacillus and Bacillus spp. are susceptible to the lytic action of Pseudomonas aeruginosa membrane vesicles. J Bacteriol. 1998;180:2306–2311. doi: 10.1128/jb.180.9.2306-2311.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadurugamuwa, J. L., and T. J. Beveridge. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other Gram-negative bacteria. Microbiology, in press. [DOI] [PubMed]
- 38.Kennedy E P. Osmotic regulation and the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:1092–1095. doi: 10.1073/pnas.79.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kessler E, Safrin M. Synthesis, processing, and transport of Pseudomonas aeruginosa elastase. J Bacteriol. 1988;170:5241–5247. doi: 10.1128/jb.170.11.5241-5247.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch A L. The biophysics of the gram-negative periplasmic space. Crit Rev Microbiol. 1998;24:23–59. doi: 10.1080/10408419891294172. [DOI] [PubMed] [Google Scholar]
- 41.Koch A L, Woeste S W. The elasticity of the sacculus of Escherichia coli. J Bacteriol. 1992;174:4811–4819. doi: 10.1128/jb.174.14.4811-4819.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo K, Takade A, Amako K. Release of outer membrane vesicles from Vibrio cholerae and Vibrio parahaemolyticus. Microbiol Immunol. 1993;37:149–152. doi: 10.1111/j.1348-0421.1993.tb03192.x. [DOI] [PubMed] [Google Scholar]
- 43.Lam J S, Graham L L, Lightfoot J, Dasgupta T, Beveridge T J. Ultrastructural examination of the lipopolysaccharides of Pseudomonas aeruginosa strains and their isogenic rough mutants by freeze-substitution. J Bacteriol. 1992;174:7159–7167. doi: 10.1128/jb.174.22.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langley S, Beveridge T J. Effect of O-side-chain-lipopolysaccharide chemistry on metal binding. Appl Environ Microbiol. 1999;65:489–498. doi: 10.1128/aem.65.2.489-498.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z, Clarke A J, Beveridge T J. A major autolysin of Pseudomonas aeruginosa, its subcellular distribution, its potential role in cell growth and division, and its secretion in surface membrane vesicles. J Bacteriol. 1996;178:2479–2488. doi: 10.1128/jb.178.9.2479-2488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Clarke A J, Beveridge T J. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J Bacteriol. 1998;180:5478–5483. doi: 10.1128/jb.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayrand D, Grenier D. Biological activities of outer membrane vesicles. Can J Microbiol. 1989;35:607–613. doi: 10.1139/m89-097. [DOI] [PubMed] [Google Scholar]
- 48.Mekalanos J J. Cholera toxin: genetic analysis, regulation and role in pathogenesis. Curr Top Microbiol Immunol. 1985;118:97–118. doi: 10.1007/978-3-642-70586-1_6. [DOI] [PubMed] [Google Scholar]
- 49.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ménard R, Prévost M-C, Gounon P, Sansonetti P J, Dehio C. The secreted Ipa complex of Shigella flexneri promotes entry into mammalian cells. Proc Natl Acad Sci USA. 1996;93:1254–1258. doi: 10.1073/pnas.93.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 52.Pesci E C, Iglewski B H. The chain of command in Pseudomonas quorum sensing. Trends Microbiol. 1997;5:132–134. doi: 10.1016/S0966-842X(97)01008-1. [DOI] [PubMed] [Google Scholar]
- 53.Pink, D., M. Jericho, and T. J. Beveridge. 1999. Unpublished data.
- 54.Plésiat P, Nikaido H. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol Microbiol. 1992;6:1323–1333. doi: 10.1111/j.1365-2958.1992.tb00853.x. [DOI] [PubMed] [Google Scholar]
- 55.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rocchetta, H. L., L. L. Burrows, and J. S. Lam. The genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev., in press. [DOI] [PMC free article] [PubMed]
- 57.Salton M R J. The relationship between the nature of the cell wall and the Gram stain. J Gen Microbiol. 1963;30:223–235. doi: 10.1099/00221287-30-2-223. [DOI] [PubMed] [Google Scholar]
- 58.Suh Y, Benedik M J. Secretion of nuclease across the outer membrane of Serratia marcescens and its energy requirements. J Bacteriol. 1997;179:677–683. doi: 10.1128/jb.179.3.677-683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wai S M, Takade A, Amako K. The release of outer membrane vesicles from the strains of enterotoxigenic Escherichia coli. Microbiol Immunol. 1995;39:451–456. doi: 10.1111/j.1348-0421.1995.tb02228.x. [DOI] [PubMed] [Google Scholar]
- 60.Whitfield C, Roberts I S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 61.Whitmire W M, Garon C F. Specific and nonspecific responses of murine B cells to membrane blebs of Borrelia burgdorferi. Infect Immun. 1993;61:1460–1467. doi: 10.1128/iai.61.4.1460-1467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wispelwey B, Hansen E J, Scheld M. Haemophilus influenzae outer membrane vesicles induced blood-brain barrier permeability during experimental meningitis. Infect Immun. 1989;57:2559–2562. doi: 10.1128/iai.57.8.2559-2562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong K R, Buckley J T. Proton motive force involved in protein transport across the outer membrane of Aeromonas salmonicida. Science. 1989;246:654–656. doi: 10.1126/science.2814486. [DOI] [PubMed] [Google Scholar]
- 64.Yao, X., M. Jericho, D. Pink, and T. J. Beveridge. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 65.Zhou L, Srisatjaluk R, Justus D E, Doyle R J. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol Lett. 1998;163:223–228. doi: 10.1111/j.1574-6968.1998.tb13049.x. [DOI] [PubMed] [Google Scholar]