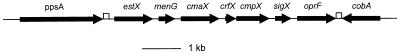
FIG. 2.
Schematic diagram of the organization of genes surrounding sigX and oprF in P. aeruginosa H103 and P. fluorescens OE 28.3. The same gene organization is observed in both species, and a combination of PCR and sequence data suggests that cmpX (putative cytoplasmic membrane protein X), sigX, and oprF are also conserved in size and gene order in P. syringae pv. syringae (data not shown). ORFs are shown as thick arrows, and the open boxes mark the locations of putative rho-independent transcription terminators. The sequence of the cobA gene in P. fluorescens has been previously reported (accession no. U09566), but the P. aeruginosa cobA gene was deduced from the Pseudomonas Genome Project sequence (20a). All other genes are putative, and similarity of cmaX, menG, estX, and ppsA was observed to an unidentified ORF from E. coli (accession no. AE000232), the E. coli S-adenosyl-methione-2-demethyl menaquinone methyl transferase gene menG, human gastric lipase, and phosphoenol pyruvate synthase, respectively. The diagram is not drawn accurately to scale.