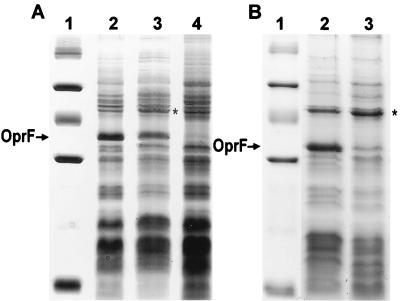
FIG. 3.
Coomassie blue-stained SDS-PAGE gel of cell envelope preparations of P. aeruginosa (A) and P. fluorescens (B) cells. (A) Lane 1, molecular weight marker (103, from top to bottom) 94, 67, 43, 30, 20.1, and 14.4; lane 2, P. aeruginosa H103 (wild type); lane 3, P. aeruginosa H814 (sigX disrupted); lane 4, P. aeruginosa H636 (oprF disrupted). (B) Lane 1, molecular weight marker (same as for panel A); lane 2, P. fluorescens OE 28.3 (wild type); lane 3, P. fluorescens FAJ2030 (sigX disrupted). The position of the band corresponding to OprF is indicated with an arrow next to both gels. Note that OprF in P. fluorescens OE 28.3 does not contain the “disulfide bridge” region (a 23-amino-acid insertion containing two disulfide bonds) found in OprF proteins in some pseudomonad strains, including P. aeruginosa H103, and so migrates further when subjected to SDS-PAGE. Note also that the cell envelope preparations shown were picked to indicate the variability in the level of reduction of OprF. The P. aeruginosa sigX mutant would frequently have a more marked reduction in OprF expression than is observed here. An asterisk marks the location of a 47-kDa protein that is increased in expression in the sigX mutant.