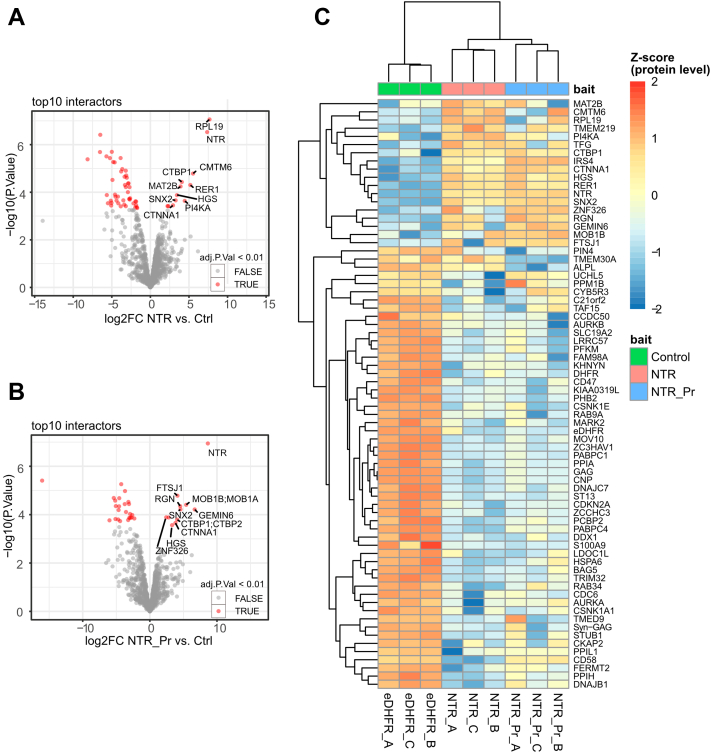
Fig. 10.
Virotrap interactome analysis of a novel protein and its N-terminal proteoform. Virotrap screens were performed in HEK293T cells using two proteoforms of the ACTB pseudogene 8 (annotated as NTR in the figures) as baits. Escherichia coli dihydrofolate reductase (eDHFR) fused to GAG was used as a negative control. A and B, volcano plots showing the interaction partners of (A) the full-length protein and (B) its N-terminal 11 amino acids shorter proteoform. Proteins with significantly altered levels are indicated in red and were defined through a pairwise t test (FDR <0.01). The x-axis shows the log2 fold change (FC) of the proteins in the ACTB pseudogene 8 Virotrap studies compared with the negative control, whereas the y-axis shows the –log10 of the adjusted p values. C, heatmap of all proteins found with significantly altered levels in at least one of the pairwise comparisons between eDHFR and the ACTB pseudogene 8 proteoforms. The scale shows Z-scored site intensity values. FDR, false discovery rate; HEK293T, human embryonic kidney 293T cell line; NTR, noncoding transcripy.