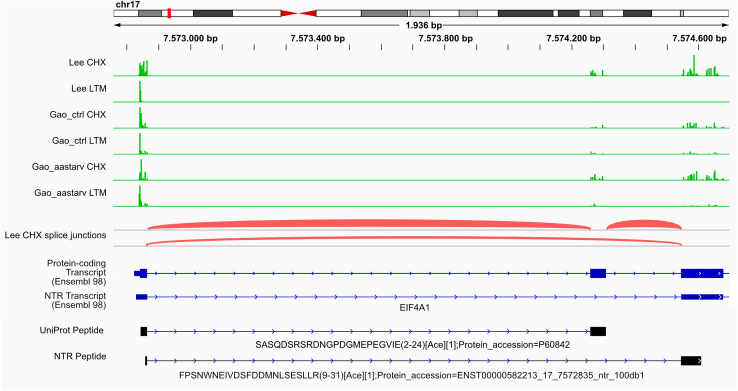
Fig. 9.
Omics evidence for an EIF4A1 proteoform visualized using a genome browser. FPSNWNEIVDSFDDMNLSESLLR peptide of the eukaryotic translation initiation factor 4A1 (EIF4A1) derived from a retained intron transcript is supported by NTR-specific alternatively spliced reads. The six top tracks represent ribosome profiling evidence of translation in Watson (green) or Crick orientation (red). We used two published studies (4, 34) as source of data for elongating ribosomes treated with cycloheximide (CHX) and initiating ribosomes treated with lactimidomycin (LTM), harvested from HEK cells under normal conditions (“Lee” and “Gao_ctrl”) or under amino acid deprivation (“Gao_aastarv”). The red tracks display the experimentally verified splice junctions. Next, transcripts (from Ensembl annotation) and genome-mapped peptides (from our study) are shown. Increasing line thickness represents introns, exons, and CDS, respectively, and arrows mark the direction of translation. The peptide name consists of peptide sequence, start–end position, N-terminal modification, spectral count, and matching protein accession. CDS, coding sequence; HEK, human embryonic kidney; NTR, noncoding transcript.