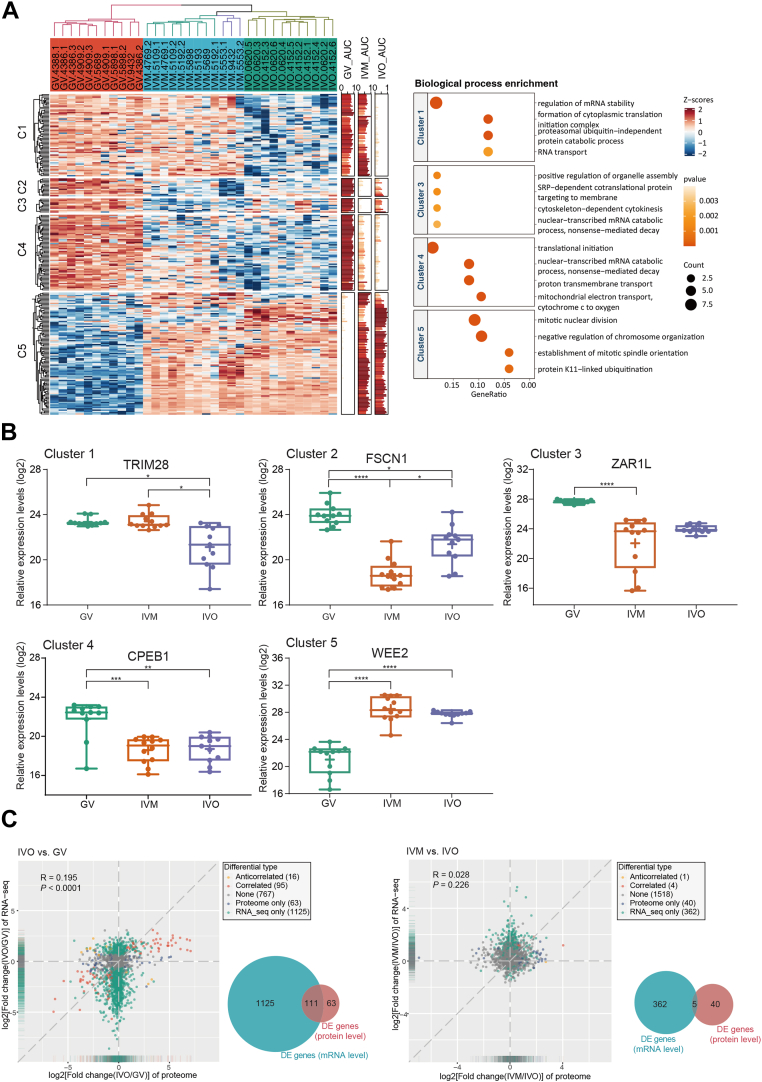
Fig. 3.
Differential protein expression analysis of human oocytes showed inconsistent mRNA and protein changes.A, heatmap (left) of 243 differentially expressed proteins among GV, IVM, IVO oocytes and enriched biological processes (right) in each cluster. Clusters of proteins (C1-C5) are indicated according to k-means. B, expression of representative proteins selected from each of the 5 clusters as interleaved boxes and whiskers with minimum and maximum values displayed. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001 (linear mixed effects model with Tukey’s posthoc test, fold change >1.5). C, scatter plot of the fold changes between the proteome and transcriptome during in vivo maturation (IVO, left) and during in vitro maturation (IVM, right) and Venn diagram of comparison of genes differentially expressed at the proteome (DE proteins) and transcriptome (DE genes) levels. Only genes identified in both transcriptome and proteome were considered. GV, germinal vesicle.