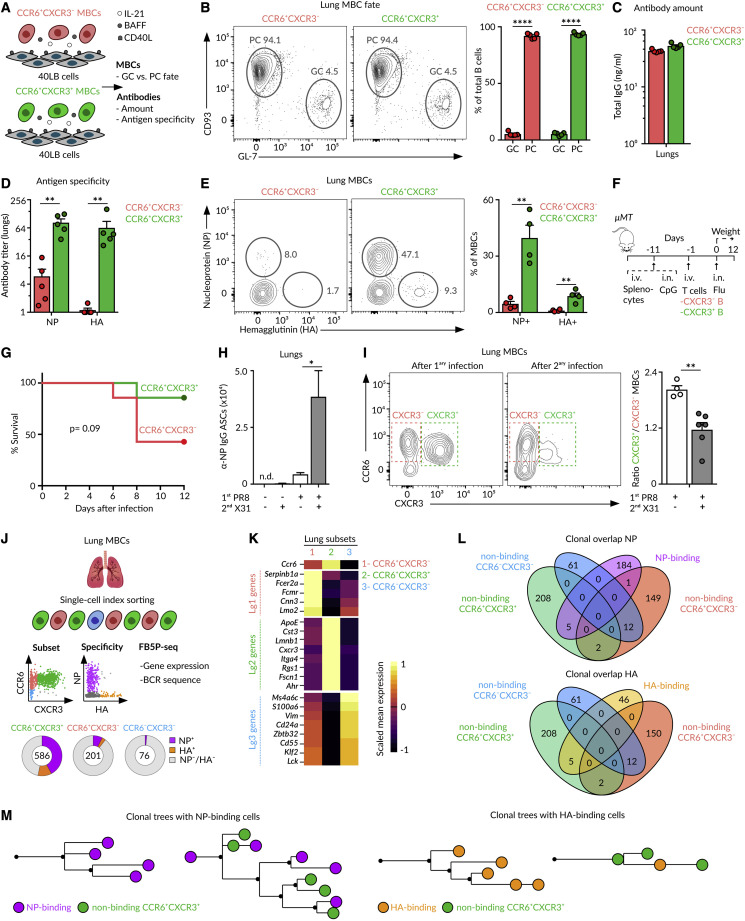
Figure 5.
MBC fate and specificity
(A) Schematic overview of the 40LB-MBCs co-culture.
(B) Contour plots showing CD93+ PCs and GL-7+ GC-like cells derived from CCR6+CXCR3− or CCR6+CXCR3+ MBCs isolated from the lungs at day 70 after the influenza infection and co-cultured for 3 days with 40LB cells.
(C and D) Bar charts displaying (C) total IgG amounts and (D) NP- or HA-specific IgG titers measured in supernatants.
(E) Contour plots showing MBC binding to NP and HA.
(F) Overview of the strategy to evaluate homotypic immunity.
(G) Survival curve of mice treated as described in (F).
(H) Enumeration of NP-specific ASCs in the lungs of mice treated with PBS or influenza PR8 and challenged at day 70 with PBS or influenza X31.
(I) Contour plots displaying MBC subsets in the lungs of mice infected with influenza PR8 alone (left panel) or re-challenged with influenza X31 after 70 days (right panel).
(J) Overview of the index cell sorting and FB5P-seq experimental workflow.
(K) Heatmap showing the expression of marker genes from Lg1, Lg2, and Lg3 clusters (10× dataset) by CCR6+CXCR3−, CCR6+CXCR3+, and CCR6−CXCR3− subsets. Color: scaled mean expression.
(L) Venn diagrams showing the clonal overlap among non-binding cells from CCR6+CXCR3−, CCR6+CXCR3+, and CCR6−CXCR3− subsets and NP-binding (upper panel) or HA-binding (lower panel) cells.
(M) Trees showing phylogenetic relationships of IgH and IgK sequences from clones containing NP-binding (left) and HA-binding (right) cells. In all quantifications: mean ± SEM, each dot represents one mouse; t test: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001.
See also Figure S4.